
Comprehensive review on the prevailing COVID-19 therapeutics and the potential of repurposing SARS-CoV-1 candidate drugs to target SARS-CoV-2 as a fast-track treatment and prevention option
Introduction
The term novel virus called severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2), has been officially designated to replacing the term human coronavirus 2019 (HCoV-19) mentioned by the International Committee on Taxonomy of viruses (1). The disagreement of surrounding the term HCoV-19 resulted in the committee’s recommendation to name as human coronavirus due to its less pathogenicity but faster spread than SARS-CoV-1 (2). The genome has already sequenced, enabling researchers to pinpoint its origin from bat through phylogenetic mapping and also identifying Pangolins as the intermediate host between Bats and Humans; but not mice or rats. COVID-19 has spread through more than 100 countries, with over 100,000 confirmed cases and 3,800 confirmed deaths worldwide at the beginning of March (3), and three month later, the high-risk global death toll has extended up to 507,435 with 10,321,689 confirmed cases as reported at the end of June by WHO (4). The rapid spread of the communicable disease, was determined based on the high reproductive number [R0] of SARS-CoV-2. The R-naught [R0] is a mathematical measure to determine the average number of people the disease has spread from a contagious person. Initially the WHO has set the R0 at 1.4–2.5 but later it was revised as being 4.7 and 6.6 R0 (5).
The crippling illness attached to COVID-19 infection have emerged as a drastic challenge to world health made worse lack of a vaccines or effective antiviral drug which could prevent or effectively cure the disease. Therefore, we aim to provide an update on drug candidates for antiviral efficacy and potent vaccines to SARS-CoV-2 and offer insights on the advantage of repurposing, drug candidates of antivirals and vaccines for SARS-CoV-1 to SARS-CoV-2. Many studies, namely in silico and in vitro, have been undertaken to develop therapeutic agents, and few have advanced up to preclinical and clinical trials on SARS-CoV-2 infections. We have placed a special focus on updating on the potent and non-potent antivirals in a nutshell for the researchers and clinicians to promote further progress in developing highly safe and potent drugs to treat COVID-19 infection successfully. More than 500 journal articles have been published since the SARS-CoV-2 outbreak, in which nearly 80 of which address the scope of effective repurposing of antivirals (3). Within the short period of outbreak, few reviews/systematic reviews have also been published (6-10). There has been more focus on the usefulness of lopinavir and chloroquine has been published (11,12). However, our review provides an update of all trialled antivirals based on their mode of action, it also emphasis their repurposing as potential anti-SARS-CoV-2 therapy. We excluded MERS or any other coronavirus species in terms of similarity of SARS-CoV-2 as only observed with SARS-CoV-1, especially in binding with the similar receptor and entry mechanism. The discussed similarity provides the possibility of repurposing drug candidates of SARS-CoV-1 to SARS-CoV-2. We present the following article in accordance with the Narrative Review reporting checklist (available at http://dx.doi.org/10.21037/atm-20-4071).
Current management of SARS-CoV-2 associated complications
When WHO declared COVID-19 disease as a global pandemic, multiple strategies on control measures such as strict physical distancing and hygienic measures were implemented by the affected countries around the world, to reduce the R° and mortality rate of SARS-CoV-2. The WHO recommended specimens such as blood cultures, upper respiratory tract, nasopharyngeal or oropharyngeal swabs were collected for RT-PCR testing where the expectorated sputum, endotracheal aspirate or bronchoalveolar lavage are later tested for positive COVID-19 infection to observe any diversified clinical symptoms (13). The most practised strategies to avoid community spread were isolation at home, quarantine measures following any travel, the lockdown on any mass gathering, and the request to maintain social distancing of 1.5 metres. The pharmacological approach depended on each country’s preference, but predominantly, chloroquine and hydroxychloroquine (HCQ) were widely used at the beginning, which has been approved by the FDA as a prophylactic drug (14,15). Within a short time, FDA has not issued the usefulness of HCQ for COVID-19 disease due to its severe side effects namely increasing heart rate, renal failure and heart attack reported in the pre-clinical trials conducted in USA. However, FDA has authorized for 800 mg intake on the first day to 50 kg or more weighed patients (16). Later, on observing the risk from large randomised clinical trials, FDA has withdrawn the approval for COVID19, however the FDA approval status for malaria, lupus and Rheumatoid arthritis will not be affected (17). China predominantly focussed on managing respiratory support and the treatment ailments as per the National Health Commission of the people’s Republic of China (18). The WHO recommended the provision of extracorporeal membrane oxygenation to critical patients with distress and refractory hypoxemia, and also high flow nasal oxygen and non-invasive ventilation has been provided for hypoxemia patients (13). In addition, endotracheal intubation was recommended for obese and pregnant patients.
Pathogenesis and clinical manifestation
Similar to SARS-CoV-1 and Middle Eastern respiratory syndrome (MERS), SARS-CoV-2 consists of structural (spike, M and nucleocapsid) and non-structural proteins (3-chymotrypsin-like protease, papain-like protease, helicase, and RNA-dependent RNA polymerase), which may act as susceptible viral targets (19). Human angiotensin-converting enzyme 2 (ACE2) is a host receptor that acts as a functional receptor and facilitates binding with the spike proteins to adopt prefusion conformational changes. Since ACE2 is expressed in many tissues such as lung, liver, heart, gastrointestinal tract and kidney, the virus can rapidly invade the body cells through this receptor and replication leads to manifest clinical symptoms (19).
COVID-19 patients show the symptoms such as fever, cough, dyspnoea, and in severe cases, may lead to the acute respiratory syndrome, pneumonia, fibrosis, renal failure, and death in high critical cases. Impaired immunity and lymphopenia are essential characteristics symptoms which upregulates, C-reactive protein (CRP) as it can be considered as significant diagnostic biomarker for COVID-19 disease (20). The predominant clinical symptom ‘pneumonia’ was observed in many patients, presented in Chest CT scan, and the abnormal features such as RNAaemia, acute respiratory distress syndrome, acute cardiac injury, and incidence of grand-glass opacities possibly lead to death as a primary endpoint (21). The incubation period of SARS-CoV-2 is 5.2 days, while 6 to 41 days is usually for the onset of critical clinical symptoms. Patients with afebrile and absence of dyspnoea may recommend for CRP count and guidance for self-isolation. The chest CT reveals patchy infiltration for positive patients (20).
Association between SARS-CoV-2 and SARS-CoV-1
Similarities and dissimilarities of SARS-CoV-1 and SARS-CoV-2
Based on genomic analysis, SARS-CoV-2 belongs to the Beta coronavirus genus of the Coronaviridae family, positive-sense, single-stranded RNA consists of 29,900 nucleotides encoding structural and non-structural proteins (22). The human coronaviruses have been studied since 1960, and the current outbreak causing SARS-CoV-2 is the seventh species of the Coronaviridae family that infect humans (23). Among these, NL-63, 229E, HKU1 and OC43 cause mild illness, while SARS-CoV-1, MERS-CoV and SARS-CoV-2 have led to severe life-threatening pandemic illness (24). Out of these three, SARS-CoV-1 and SARS-CoV-2 share the same entry mechanism and receptor called ACE-2, which is an essential residue for the binding site of these viruses, whereas MERS-CoV binds to the receptor dipeptidyl peptidase 4 (25). Since SARS-CoV-1 and SARS-CoV-2 bind to the same human receptor, viral entry could be considered as an essential target for predicting the utility of currently used antivirals. The spike protein is the main key for entry into host-cell through ACEII receptor on priming with other co-factors. Therefore, effective SARS-CoV-1 therapies targeted to the entry site could be explored for use in identifying potent antivirals to inhibit SARS-CoV-2 multiplication.
Homology modelling demonstrates 80% similarity between SARS-CoV-1 and SARS-CoV-2 (26). In particularly the spike protein of SARS-CoV-1 and SARS-CoV-2 shares 75% similarity towards amino acid sequence (27). The SARS-CoV-2 binds with stronger affinity to the ACE receptor than SARS-CoV-1, with maximum proximal similar amino acid residues (22). The ACE receptor is membrane-associated aminopeptidase that consists of a high level of viral regulatory genes which are involved in the viral life cycle. Figure 1 represents the mechanism of entry and possible targets for SARS-CoV-2.
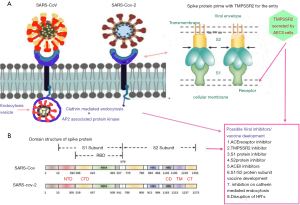
Grifoni et al. compared the protein sequence through a homology modelling and bioinformatics approach. They demonstrated that SARS-CoV-1, MERS-CoV and bat-SL-CoVZXC21 revealed the similarities with SARS-CoV-2 as 76%, 35% and 80% of the spike protein respectively (28). Among the different strains, bat-SL-CoZXC21, non-human strain was showed highest similarity. SARS-CoV-1 spike protein has expressed as highest similarities than MERS-CoV. The authors have reported the B cell epitopes sequencing of SARS-CoV-2 and SARS-CoV-1 regions from immune epitope databases and studied the Spike proteins similarities, where the resemblances were ranging from 69–100%. In addition the study has reported sequencing ORF1ab (86%), E (94%), M (90%), N (90%), S (76%) to find the protein similarities of SARS-CoV-1 with MERS-CoV as ORF1ab (50%), E (36%), M (42%), N (48%), S (45%). In addition, the study compared B and T cell immunodominant SARS-CoV-1 was mapped to the homologous SARS-CoV-2 proteins and found to have a high percentage of similarity (28).
Kumar et al. studied the spike protein sequence of SARS-CoV-2 with SARS-CoV-1 using EMBOSS Needle pairwise sequence alignment tools (29). The authors revealed that 12.8% of difference observed in the S protein between both strains, especially in the amino acid sequence alignment. In addition, 83.9% similarity was observed in the minimal receptor-binding domain (RBD) with SARS-CoV-1. The RBD of SARS-CoV-2 has 73% similarity with SARS-CoV-1 RBD (30). The phylogenetic analysis study has revealed the overall sequence similarities for SARS-CoV-1 and SARS-CoV-2 for around 76–78% for the whole protein, 73–76% for the RBD, and 50–53% for the RBM (22). The Sequence alignment between SARS-CoV-1 (RBD219-N1) and SARS-CoV-2 spike protein has shown 75% identity and 83% similarity in RBD region (31). Therefore, based on these similarities, our review has focussed on the antivirals used to treat SARS-CoV-1 that could be considered for use against SARS-CoV-2.
The significant dissimilarities between these SARS-CoV-1 and SARS-CoV-2 occur in the genomic material, non-structural proteins and in nucleocapsid. The open reading frame of SARS-CoV-2 was sequenced and compared with the SARS-CoV-1 viral genome, and a mutation was observed in the non-structural proteins (NS2 and NS3). The observed destabilising mutation may be the reason for the rapid spread and seriousness of the infection (32). The comprehensive support on the difference between these two strains has been observed in non-structural and accessory proteins. They are similar in non-structural protein alignment but vary in structural amino acids. For instance, SARS-CoV-2 does not contain 8a protein but has a long amino acid chain with 121 amino acids, which is not in SARS-CoV-1, and a similar variation occurs in the 3b protein (33). Therefore, our review will provide clear insights to focus more on the spike protein and entry-level mechanism of SARS-CoV-2 for potent antivirals and vaccines development.
Repurposing of SARS-CoV-1 entry-inhibitors other than the tested antivirals for COVID-19
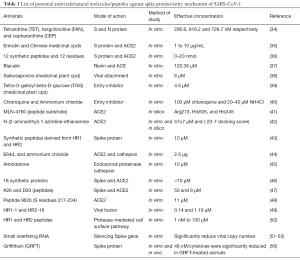
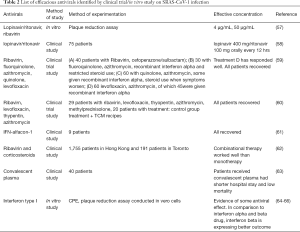
Repurposing antivirals from amongst viral strains is an intelligent strategy to find out the effective antivirals against contagious novel pandemic strain like SARS-CoV-2. Therefore, our study has mainly attributed the similarities between the SARS-CoV-1 and SARS-CoV-2 as they both share significant similarities, including beta-genus family and extends the similarities in spike protein, entry mechanism and RBD as shown in the Figure. However, the difference exists in few amino acid sequences aligned in spike protein; the domain arrangements of similarity exist to SARS-CoV-1 to SARS-CoV-2. Hence our study also recommends for repurposing the antivirals which were potential to spike protein inhibition against SARS-CoV-1, either in vitro or in silico evaluation were highlighted in Table 1 (34-56) as these antivirals were not studied on SARS-CoV-2 so far. Table 2 recommends a few efficient non target based antivirals against SARS-CoV-1 either through in vitro or clinical studies has been highlighted for future trial purpose (57-66). Shanmugaraj et al. has summarised the therapeutic measures of potential monoclonal antibody against SARS-CoV-1 and MERS-CoV entry mechanism as a significant target where the current review has focussed on antivirals exclusively (25). Haagmans et al. has summarised possible in vitro and in vivo neutralising monoclonal antibodies, and potential immunotherapy against SARS-CoV-1 entry inhibitors has been updated (67).
Full table
Full table
Update on current antiviral therapy on SARS-CoV-2
Drug discovery against SARS-CoV-2 by in silico study
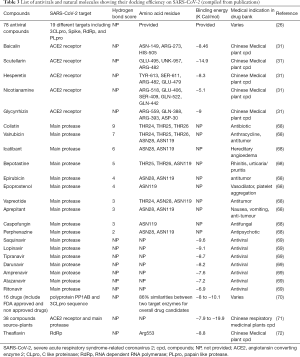
The computational approach to screen the effective antivirals from the available compounds is much desirable, time consuming and more rapid method to screen the potent compound when compared to in vitro and in vivo evaluation. Within the short period, several research crews have studied the molecular docking with available proteins from the PDB and docked with a compound ligand structure. Table 3 explicits the efficiently docked chemical compounds to various protein targets of SARS-CoV-2. Many of the docking studies have attempted with the main protease of SARS-CoV-2 using online docking software. There are seven in silico studies investigated the binding affinity against SARS-CoV-2 proteins and enzymes (26,31,68-72). Saquinavir (SQV) and lopinavir binding energy with the main protease was higher than HIV and SARS-CoV-1 (69). Wu et al. have studied 78 different antivirals on various SARS-CoV-2 protein (26). Remdesivir has exhibited high binding energy on Nsp3b (–36.5), RdRp (–112.8) amino acid residues. Darunavir has docked to NsP3c (–126.149), PLpro (–110.759) as chloroquine has docked to Nsp3b with the binding energy (–130.355) (26). Disappointingly darunavir has not shown any potential activity against SARS-CoV-2 strain through in vitro study at the concentration of EC50 >100 µM which was compared with remdesivir as a positive control (73).
Full table
Drug discovery by in vitro study against SARS-CoV-2
The effective drug discovery approach by in vitro study from the existing drug is the second fastest protocol to identify antivirals against newly emerged viral infections. Wang et al. have studied multiple drugs including remdesivir, chloroquine (GS5734), ribavirin, penciclovir, nitazoxanide, nafamostat, chloroquine and favipiravir against SASR-CoV-2 (74). Among these, remdesivir (EC50 =0.77 µM) and chloroquine (EC50 =1.13) are identified as a potent inhibitor against SARS-CoV-2 at low molar concentration. Evidently, in time dose-dependent assay chloroquine has shown appreciable inhibitory activity on before and after entry level experimentation. Lopinavir/Ritonavir has a significant role in SARS-CoV-2 and so where tested in 16 patients reported in Wuhan city either independently or in the combination of arbidol (75). The investigation supports that combinational treatment has been more efficacious than lopinavir/ritonavir as monotherapy. As per the Chinese national health guidelines, lopinavir/ritonavir (400 mg/100 mg bid po) and IFN-alpha (5 million U bid inh) are recommended for SARS-CoV-2 treatment (18,76).
HCQ (EC50 =0.72 µM) was found more potent against SARS-CoV-2 than chloroquine which exhibited as EC50 =5.7 µM in Vero cell line (77). Ivermectin (5 µM) has exhibited potential inhibitory activity against SARS-CoV-2 at 5,000 fold reduction of viral copy number compared to the control and IC50 was around approximately 2 µM based on protein target dependence of infected strain (78). Lianhuaqingwen (LH), a compound from the Chinese medicinal plant, was evaluated for its efficacy against SARS-CoV-2 showing an IC50 411.2 µg/mL (LH) by CPE assay and considering remdesivir, as positive control at IC50 of 0.651 µM by plaque reduction assay (79). The authors have also studied the immunomodulatory effect of LH and observed elevated expression of four cytokines (TNF-α, IL-6, CCL-2/MCP-1, and CXCL-10/IP-10) with a significant difference. The antiviral activity of ribonucleoside analogue ßd-N4-hydroxycytidine (NHC) (EIDD-1931) was studied against SARS-CoV-2, SARS-CoV-1 and MERS-CoV in Vero cells and Calu-3 cells (80). The effective inhibitory concentration was IC50 of 0.3 µM and CC50 of >10 µM in Vero cells and 0.08 µM in Calu-3 cells.
Hoffmann et al. stated that the spike protein shares about 76% amino acid identity with SARS-CoV-1 (81) and studied the mechanism of entry through ACE receptor to the host cell. The research crew has performed in vitro study and demonstrated the cellular serine protease, TMPRSS2 priming with SARS-CoV-2 for the entry. The study demonstrated the Camostat mesylate as TMPRSS2 inhibitor which blocks the activity of CatB/L and TMPRSS2 priming and thereby inhibits entry of SARS-CoV-2 virus into host cell (81). Teicoplanin has shown a better antiviral efficiency on SARS-CoV-2 at 1.66 µM 50% inhibitory concentration against targeting cathepsin L protein which is used for cell entry where vancomycin and other test antibiotics did not exhibit any appreciable inhibitory activity against Cathepsin L protein (82).
Highlights from clinicians for medications against COVID-19 through publications
A case report on US first patient’s medications history could help other clinical practice to treat baseline respiratory syndrome symptoms (83). Once after the confirmation of positive PCR test results, the patient had been treated according to his/her symptoms existed. Apart from the antipyretic and hospital-acquired pneumonia treatment, the patient had treated with remdesivir and provided with oxygen supply (83). Zhang et al. have mentioned oral moxifloxacin or levofloxacin (consider tolerance) and arbidol for COVID-19 bacterial co-infection treatment (20). The emergency cases, were treated with antiviral plus anti-pneumococcus plus antiStaphylococcus aureus with nemonoxacin (750 mg once daily) and linezolid (20). SpO2 <90%, dexamethasone 5–10 mg or methylprednisolone 40–80 mg was given intravenously for emergency cases. Oral oseltamivir was widely used in china hospitals for the treatment for COVID-19 cases which is considered as neuraminidase inhibitors (76). Corticosteroids are not recommended for COVID-19 disease where it has shown numerous side effects such as septic shock, myocardial lung injury and similar symptoms of acute respiratory syndrome disease (84). However, based on the china government guidelines, some clinicians are recommending corticosteroids in a mild dose (≤0.5–1 mg/kg per day methylprednisolone or equivalent) for ≤7 days to treat critical cases (85).
The synergistic effect of the combinational treatment of HCQ (600 mg/d for 10 days) and azithromycin (500 mg day 1 and 250 mg days 2 to 5) has treated for 36 COVID-19 patients based on the PCR confirmation. Among the selected cohorts, 26 patients had received HCQ and 10 were in the control group (86). Among these 26, six patients have showed 100% cure who has administered with HCQ and azithromycin whereas drug HCQ alone has shown 57% cure from COVID-19 disease. Perhaps, none of the patients has received azithromycin alone. Hence it would be appreciated to call it combinational efficacy than its synergistic efficacy. Similarly, the synergistic effect has reported by the same clinical team from France in 80 clinically ill patients (includes six patients published in previous publication) on the potential treatment of HCQ and azithromycin (87). All patients have responded well with the synergistic treatment except two non-responders. However, the disagreement on using of HCQ and Azithromycin combination has been raised in the study which is the highlights to be considered. The prospective study has administered HCQ (600 mg/d for 10 days) and azithromycin (500 mg day 1 and 250 mg days 2 to 5) conducted in 11 consecutive patients. The results were still observed the positive viral load even on 6th day after the combinational treatment initiation (88).
The monotherapy of HCQ have demonstrated effective and appreciable results in few studies which should be noted. In a prospective study, 30 patients have received 400 mg of HCQ per day for five consecutive days plus conventional treatments and were compared with a group who received only conventional treatment (89). On day 7, the effect of HCQ was observed in 13 (86.7%) cases where the control group had observed 14 (93.3%) cases on evidence as negative in throat swabs. The effect of HCQ was almost equal and effective as conventional treatment, and hence authors suggested to study with a large cohort to investigate the effect of HCQ in COVID19 patients (89). Yao et al. have suggested HCQ (400 mg) can be given twice daily/day and gradually 200 mg of HCQ twice daily for four more days as a maintenance dose for the COVID19 treatment (77). The research crew has stated HCQ (EC50 =0.72 µM) was efficient against SARS-CoV-2, than chloroquine (EC50 =5.47 µM) and also studied the plasma/blood profile through pharmacokinetics by in silico methods. Comparatively blood and plasma concentration of HCQ have been increased rapidly and maintained steady stability (77). Though the drug remdesivir has demonstrated side effects in 102 (66%) of 155 remdesivir recipients, it was reported as an effective therapeutic agent on developing appreciable clinical improvement when compared to placebo (90).
Immunotherapy
Antibody therapy and immunotherapy is another genre of therapeutic measures. The following segment summarises the published domains attempt for any invention. The possible cross-protection of SARS-CoV-1 RBD against SARS-CoV-2 can be a potential target as earlier results of anti-SARS vaccines have cross neutralise other Bat-originated SARS strains (91). SARS-CoV-1 potent neutralising antibodies m396, CR3014, CR3022 were tested for the efficacy on comparing with irrelevant anti-CD40 as control. The anti-SARS-CoV-1 RBD neutralising mAbs 80R and S230 were studied for the docking affinity to SARS-CoV-2 proteins by virtual screening (92). Among these CR3022 showed better affinity towards ACE2 binding site of SARS-CoV-2 while the other two antibodies failed to bind with SARS-CoV-2 spike protein (30). Based on the study results, CR3022, could be considered as significant therapeutic measures or vaccine candidates for COVID-19 disease. Grifoni et al. have studied the SARS-CoV-1 B cell and T cell epitope mapped with SARS-CoV-2; here-in demonstrated that they both have an average 60–90% similarity (28). Recently convalescent plasma therapy is a classic adaptive immunotherapy to consider ailments for many diseases. Likewise, plasma therapy is contributing its importance as a promising treatment option to COVID-19 disease indeed. Clinicians has treated on 10 severe patients with one dose of 200 mL of convalescent Plasma transfusion with neutralising antibody titre 1:640 and observed massive development in median time 16.5 days where there were no any severe adverse effects which is noticeable (93). Passive immunisation is the best effective prophylaxis of any viral infection. Convalescent plasma (IgG) with a binding titre greater than 1:1,000 dilution was transfused to five critically ill patients and observed better improvement in all the patients (94).
Update on antivirals registered for clinical trials
As of April 11, a total of 51 recruited clinical trials has been started aiming to evaluate the efficacy of antiviral for the treatment of COVID-19 infections all over the world, as recorded in NIH, US library of Medicine (https://clinicaltrials.gov/ct2/home) is tabulated (Table 4). Indian clinical trials committee has launched clinical trials on HCQ against SARS-CoV-2 (CTRI/2020/03/024402) (Table 4). China has launched 303 ongoing clinical trials, among them, 16.5% (50 trials) has attempting with compounds from traditional Chinese Medicine and 4.6% (14 trials) were analysed for the combined effect of TCM with Western Medicine (10). The Chinese clinical trials are available on Chinese clinical trial registry (http://www.chictr.org.cn/abouten.aspx).
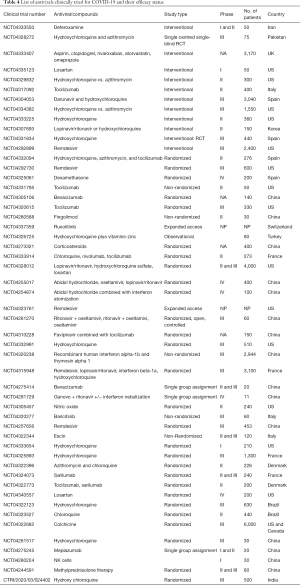
Full table
Strategies on vaccines development for SARS-CoV-2
The S protein plays a significant role in neutralising antibody, T cell response and defensive immunity developed by their viruses during the infection (95). There are different types of target available for vaccines development such as full-length S protein, DNA-based, Viral vector, recombinant S protein-based, RBD, DNA-based, viral vector-based, Recombinant RBD protein-based development. Each of the mentioned vaccine candidate has their advantage and disadvantages on respective strategy (95). Only a small number of vaccine development has attempted for SARS-CoV-1 which had reached clinical trials. WHO reports about 120 projects from many pharmaceutical companies and universities on the development of a vaccines have been registered all over the world (96). Among that, six had been approved for clinical trials for evaluation.
The American company, Moderna has registered for phase I clinical trial on mRNA vaccine development (phase 1 clinical trial NCT04283461). Innovio has started on DNA vaccine development with 40 volunteers (phase 1 NCT04336410) started at the beginning of April. The University of Queensland had started to work on the virus in cell cultures which are at preclinical testing (which are hopefully started at early April). The University of Oxford in England has started recombinant vaccine trials in 500 volunteers (phase 1/2 NCT04324606). Johnson and Johnson and Sanofi are joining hands for the vaccine development to SARS-CoV-2 with 200 volunteers. Inactivated candidate vaccine has been approved from the developer Sinovac and Beijing Institute of Biological Products/Wuhan Institute of Biological Products starting from April (phase 2 ChiCTR2000031781 and phase 1 ChiCTR2000030906). Meanwhile, some vaccines development crew from Netherland and Australia are aiming to conduct clinical trials on the use of tuberculosis vaccine to SARS-CoV-2.
Summary and perspectives
The rapid pandemic swept of COVID-19 disease across China, and the world made it as a global health crisis recently. Since the elderly and medically critically ill patients are more susceptible, it requires effective instant antivirals to be identified as soon as possible. The pharmaceutical industry and clinical trials are working to develop antivirals to combat COVID-19 disease. Thanks to research crews for characterisation of viral life cycle and studies on viral characteristics within the short period that could be able to bring out the similarities and dissimilarities with SARS-CoV-1 that predict several hosts and viral proteins targeting molecules to provide the promising antiviral candidates. We have examined published evidence in support of the similarities between SARS-CoV-2 and SARS-CoV-1 in entry-level mechanism and spike protein alignment that enable us to predict the effective antivirals. Owing to the major appreciable similarities in structure and clinical manifestation, leaving aside high R°, the most efficacious drug on SARS-CoV-1 infection could be tried as antivirals to treat COVID19. Also, the expertise and potent highly immunogenic/antiviral component identified for SARS-CoV-1 would be consider for repurposing against spike-based or entry-based target to inhibit SARS-CoV-2 replication. Our review might galvanize the current research and professional community to evolve with significant findings to treat COVID-19 disease.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-4071
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at available at http://dx.doi.org/10.21037/atm-20-4071). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Gorbalenya AE, Baker SC, Baric RS, et al. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 2020;5:536-44. [Crossref] [PubMed]
- Wu Y, Ho W, Huang Y, et al. SARS-CoV-2 is an appropriate name for the new coronavirus. Lancet 2020;395:949-50. [Crossref] [PubMed]
- Liu C, Zhou Q, Li Y, et al. Research and Development on Therapeutic Agents and Vaccines for COVID-19 and Related Human Coronavirus Diseases. ACS central science 2020;6:315-31. [Crossref] [PubMed]
- Coronavirus death report - WHO [database on the Internet] 2020. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019. Accessed: 30-6-2020.
- Sanche S, Lin YT, Xu C, et al. The novel coronavirus, 2019-nCoV, is highly contagious and more infectious than initially estimated. arXiv preprint arXiv:200203268 2020.
- Wang L, Wang Y, Ye D, et al. A review of the 2019 Novel Coronavirus (COVID-19) based on current evidence. Int J Antimicrob Agents 2020;56:10613. [Crossref] [PubMed]
- Guo YR, Cao QD, Hong ZS, et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil Med Res 2020;7:11. [PubMed]
- Zhang L, Liu Y. Potential interventions for novel coronavirus in China: A systematic review. J Med Virol 2020;92:479-90. [Crossref] [PubMed]
- Pang J, Wang MX, Ang IYH, et al. Potential Rapid Diagnostics, Vaccine and Therapeutics for 2019 Novel Coronavirus (2019-nCoV): A Systematic Review. J Clin Med 2020.9. [PubMed]
- Yang Y, Islam MS, Wang J, et al. Traditional Chinese Medicine in the Treatment of Patients Infected with 2019-New Coronavirus (SARS-CoV-2): A Review and Perspective. Int J Biol sci 2020;16:1708-17. [Crossref] [PubMed]
- Cortegiani A, Ingoglia G, Ippolito M, et al. A systematic review on the efficacy and safety of chloroquine for the treatment of COVID-19. J Crit Care 2020;57:279-83. [Crossref] [PubMed]
- Yao TT, Qian JD, Zhu WY, et al. A systematic review of lopinavir therapy for SARS coronavirus and MERS coronavirus-A possible reference for coronavirus disease-19 treatment option. J Med Virol 2020;92:556-63. [Crossref] [PubMed]
- WHO. Clinical management of severe acute respiratory infection when novel coronavirus (nCoV) infection is suspected. Available online: https://www.who.int/publications-detail/clinical-management-of-severe-acute-respiratory-infection-when-novel-coronavirus-(ncov)-infection-is-suspected
- Lenzer J. Covid-19: US gives emergency approval to hydroxychloroquine despite lack of evidence. BMJ 2020;369:m1335. [Crossref] [PubMed]
- Ferner RE, Aronson JK. Chloroquine and hydroxychloroquine in covid-19. BMJ 2020;369:m1432. [Crossref] [PubMed]
- . .Hydroxychloroquine FDA. 2020.
- Coronavirus (COVID-19) Update: FDA Revokes Emergency Use Authorization for Chloroquine and Hydroxychloroquine [database on the Internet]2020. Available online: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-revokes-emergency-use-authorization-chloroquine-and. Accessed: 2-6-2020
- National Health Commission of the People's Republic of China. [database on the Internet]. Notice on printing and distributing the diagnosis and treatment plan of pneumonia with new coronavirus infection (trial version 3). Available online: http://www.nhc.gov.cn/yzygj/s7653p/202001/f492c9153ea9437bb587ce2ffcbee1fa.shtml. Accessed: 8 April 2020.
- Astuti I. Ysrafil. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): An overview of viral structure and host response. Diabetes Metab Syndr 2020;14:407-12. [Crossref] [PubMed]
- Zhang J, Zhou L, Yang Y, et al. Therapeutic and triage strategies for 2019 novel coronavirus disease in fever clinics. Lancet Respir Med 2020;8:e11-2. [Crossref] [PubMed]
- Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497-506. [Crossref] [PubMed]
- Wan Y, Shang J, Graham R, et al. Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J Virol 2020;94:e00127-20. [Crossref] [PubMed]
- Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 2019;17:181-92. [PubMed]
- Zhu N, Zhang D, Wang W, et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med 2020;382:727-33. [Crossref] [PubMed]
- Shanmugaraj B, Siriwattananon K, Wangkanont K, et al. Perspectives on monoclonal antibody therapy as potential therapeutic intervention for Coronavirus disease-19 (COVID-19). Asian Pac J Allergy Immunol 2020;38:10-8. [PubMed]
- Wu C, Liu Y, Yang Y, et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica B 2020;10:766-88. [Crossref] [PubMed]
- Xu X, Chen P, Wang J, et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci 2020;63:457-60. [Crossref] [PubMed]
- Grifoni A, Sidney J, Zhang Y, et al. A Sequence Homology and Bioinformatic Approach Can Predict Candidate Targets for Immune Responses to SARS-CoV-2. Cell Host Microbe 2020;27:671-680.e2. [Crossref] [PubMed]
- Kumar S, Maurya VK, Prasad AK, et al. Structural, glycosylation and antigenic variation between 2019 novel coronavirus (2019-nCoV) and SARS coronavirus (SARS-CoV). Virusdisease 2020;31:13-21. [Crossref] [PubMed]
- Tian X, Li C, Huang A, et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect 2020;9:382-5. [Crossref] [PubMed]
- Chen H, Du Q. Potential natural compounds for preventing 2019-nCoV infection. Preprints 2020:202001.0358.v1.
- Angeletti S, Benvenuto D, Bianchi M, et al. COVID-2019: The role of the nsp2 and nsp3 in its pathogenesis. J Med virol 2020;92:584-8. [Crossref] [PubMed]
- Wu A, Peng Y, Huang B, et al. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe 2020;27:325-8. [Crossref] [PubMed]
- Kim DE, Min JS, Jang MS, et al. Natural Bis-Benzylisoquinoline Alkaloids-Tetrandrine, Fangchinoline, and Cepharanthine, Inhibit Human Coronavirus OC43 Infection of MRC-5 Human Lung Cells. Biomolecules 2019;9:696. [Crossref] [PubMed]
- Ho TY, Wu SL, Chen JC, et al. Emodin blocks the SARS coronavirus spike protein and angiotensin-converting enzyme 2 interaction. Antivir Res 2007;74:92-101. [Crossref] [PubMed]
- Ho TY, Wu SL, Chen JC, et al. Design and biological activities of novel inhibitory peptides for SARS-CoV spike protein and angiotensin-converting enzyme 2 interaction. Antivir Res 2006;69:70-6. [Crossref] [PubMed]
- Deng YF, Aluko RE, Jin Q, et al. Inhibitory activities of baicalin against renin and angiotensin-converting enzyme. Pharm Biol 2012;50:401-6. [Crossref] [PubMed]
- Cheng PW, Ng LT, Chiang LC, et al. Antiviral effects of saikosaponins on human coronavirus 229E in vitro. Clin Exp Pharmacol Physiol 2006;33:612-6. [Crossref] [PubMed]
- Yi L, Li Z, Yuan K, et al. Small molecules blocking the entry of severe acute respiratory syndrome coronavirus into host cells. J Virol 2004;78:11334-9. [Crossref] [PubMed]
- Vincent MJ, Bergeron E, Benjannet S, et al. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol J 2005;2:69. [Crossref] [PubMed]
- Towler P, Staker B, Prasad SG, et al. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J Biol Chem 2004;279:17996-8007. [Crossref] [PubMed]
- Huentelman MJ, Zubcevic J, Prada JAH, et al. Structure-Based Discovery of a Novel Angiotensin-Converting Enzyme 2 Inhibitor. Hypertension 2004;44:903-6. [Crossref] [PubMed]
- Liu S, Xiao G, Chen Y, et al. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet 2004;363:938-47. [Crossref] [PubMed]
- Huang IC, Bosch BJ, Li F, et al. SARS Coronavirus, but Not Human Coronavirus NL63, Utilizes Cathepsin L to Infect ACE2-expressing Cells. J Biol Chem 2006;281:3198-203. [Crossref] [PubMed]
- Stadler K, Ha HR, Ciminale V, et al. Amiodarone Alters Late Endosomes and Inhibits SARS Coronavirus Infection at a Post-Endosomal Level. Am J Respir Cell Mol Biol 2008;39:142-9. [Crossref] [PubMed]
- Kao RY, Tsui WH, Lee TS, et al. Identification of novel small-molecule inhibitors of severe acute respiratory syndrome-associated coronavirus by chemical genetics. Chem Biol 2004;11:1293-9. [Crossref] [PubMed]
- Han DP, Penn-Nicholson A, Cho MW. Identification of critical determinants on ACE2 for SARS-CoV entry and development of a potent entry inhibitor. Virology 2006;350:15-25. [Crossref] [PubMed]
- Guo Y, Tisoncik J, McReynolds S, et al. Identification of a new region of SARS-CoV S protein critical for viral entry. J Mol Biol 2009;394:600-5. [Crossref] [PubMed]
- Yuan K, Yi L, Chen J, et al. Suppression of SARS-CoV entry by peptides corresponding to heptad regions on spike glycoprotein. Biochem Biophys Res Commun 2004;319:746-52. [Crossref] [PubMed]
- Ujike M, Nishikawa H, Otaka A, et al. Heptad Repeat-Derived Peptides Block Protease-Mediated Direct Entry from the Cell Surface of Severe Acute Respiratory Syndrome Coronavirus but Not Entry via the Endosomal Pathway. J Virol 2008;82:588-92. [Crossref] [PubMed]
- Akerström S, Mirazimi A, Tan YJ. Inhibition of SARS-CoV replication cycle by small interference RNAs silencing specific SARS proteins, 7a/7b, 3a/3b and S. Antiviral Res 2007;73:219-27. [Crossref] [PubMed]
- He ML, Zheng BJ, Chen Y, et al. Kinetics and synergistic effects of siRNAs targeting structural and replicase genes of SARS-associated coronavirus. FEBS Lett 2006;580:2414-20. [Crossref] [PubMed]
- Zheng BJ, Guan Y, Tang Q, et al. Prophylactic and therapeutic effects of small interfering RNA targeting SARS-coronavirus. Antivir Ther 2004;9:365-74. [PubMed]
- Qin ZL, Zhao P, Zhang XL, et al. Silencing of SARS-CoV spike gene by small interfering RNA in HEK 293T cells. Biochem Biophys Res Commun 2004;324:1186-93. [Crossref] [PubMed]
- Zhang Y, Li T, Fu L, et al. Silencing SARS-CoV Spike protein expression in cultured cells by RNA interference. FEBS Lett 2004;560:141-6. [Crossref] [PubMed]
- O'Keefe BR, Giomarelli B, Barnard DL, et al. Broad-spectrum in vitro activity and in vivo efficacy of the antiviral protein griffithsin against emerging viruses of the family Coronaviridae. J Virol 2010;84:2511-21. [Crossref] [PubMed]
- Chu CM, Cheng VCC, Hung IFN, et al. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax 2004;59:252-6. [Crossref] [PubMed]
- Chan KS, Lai ST, Chu CM, et al. Treatment of severe acute respiratory syndrome with lopinavir/ritonavir: a multicentre retrospective matched cohort study. Hong Kong Med J 2003;9:399-406. [PubMed]
- Zhao Z, Zhang F, Xu M, et al. Description and clinical treatment of an early outbreak of severe acute respiratory syndrome (SARS) in Guangzhou, PR China. J Med Microbiol 2003;52:715-20. [Crossref] [PubMed]
- Qiang J, Biao W, Rui-lin Z, et al. Clinical controlled study of integrative Chinese and western medicine in treating 49 cases of SARS. Chin J Integr Med 2003;9:175-80. [Crossref]
- Loutfy MR, Blatt LM, Siminovitch KA, et al. Interferon alfacon-1 plus corticosteroids in severe acute respiratory syndrome: a preliminary study. JAMA 2003;290:3222-8. [Crossref] [PubMed]
- Lau EHY, Cowling BJ, Muller MP, et al. Effectiveness of ribavirin and corticosteroids for severe acute respiratory syndrome. Am J Med 2009;122:1150.e11-21. [Crossref] [PubMed]
- Soo YO, Cheng Y, Wong R, et al. Retrospective comparison of convalescent plasma with continuing high-dose methylprednisolone treatment in SARS patients. Clin Microbiol Infect 2004;10:676-8. [Crossref] [PubMed]
- Chen F, Chan KH, Jiang Y, et al. In vitro susceptibility of 10 clinical isolates of SARS coronavirus to selected antiviral compounds. J Clin Virol 2004;31:69-75. [Crossref] [PubMed]
- Cinatl J, Morgenstern B, Bauer G, et al. Treatment of SARS with human interferons. Lancet 2003;362:293-4. [Crossref] [PubMed]
- Scagnolari C, Vicenzi E, Bellomi F, et al. Increased sensitivity of SARS-coronavirus to a combination of human type I and type II interferons. Antivir Ther 2004;9:1003-11. [PubMed]
- Haagmans BL, Osterhaus AD. Coronaviruses and their therapy. Antivir Res 2006;71:397-403. [Crossref] [PubMed]
- Liu X, Wang XJ. Potential inhibitors against 2019-nCoV coronavirus M protease from clinically approved medicines. J Genet Genomics 2020;47:119-21. [Crossref] [PubMed]
- Ortega JT, Serrano ML, Pujol FH, et al. Unrevealing sequence and structural features of novel coronavirus using in silico approaches: The main protease as molecular target. EXCLI Journal 2020;19:400-9. [PubMed]
- Chen YW, Yiu CPB, Wong KY. Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CL (pro)) structure: virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Research 2020;9:129. [Crossref] [PubMed]
- Yan YS, Cao Y, Zhang J, et al. Discovery of Anti-2019-nCoV Agents from Chinese Patent Drugs via Docking Screening. Preprints 2020;2020020254:2020.
- Lung J, Lin YS, Yang YH, et al. The potential chemical structure of anti-SARS-CoV-2 RNA-dependent RNA polymerase. J Med Virol 2020;92:693-7. [Crossref] [PubMed]
- De Meyer S, Bojkova D, Cinati J, et al. Lack of Antiviral Activity of Darunavir against SARS-CoV-2. medRxiv 2020:2020.04.03.20052548.
- Wang M, Cao R, Zhang L, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 2020;30:269-71. [Crossref] [PubMed]
- Deng L, Li C, Zeng Q, et al. Arbidol combined with LPV/r versus LPV/r alone against Corona Virus Disease 2019: A retrospective cohort study. J Infect 2020;81:e1-5. [Crossref] [PubMed]
- Lu H. Drug treatment options for the 2019-new coronavirus (2019-nCoV). Biosci Trends 2020;14:69-71. [Crossref] [PubMed]
- Yao X, Ye F, Zhang M, et al. In Vitro Antiviral Activity and Projection of Optimized Dosing Design of Hydroxychloroquine for the Treatment of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Clin Infect Dis 2020;71:732-9. [Crossref] [PubMed]
- Caly L, Druce JD, Catton MG, et al. The FDA-approved Drug Ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res 2020;178:104787. [Crossref] [PubMed]
- Runfeng L, Yunlong H, Jicheng H, et al. Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2). Pharmacol Res 2020;156:104761. [Crossref] [PubMed]
- Sheahan TP, Sims AC, Zhou S, et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci Transl Med 2020;12:eabb5883.
- Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020;181:271-280.e8. [Crossref] [PubMed]
- Zhang J, Ma X, Yu F, et al. Teicoplanin potently blocks the cell entry of 2019-nCoV. bioRxiv 2020:2020.02.05.935387.
- Holshue ML, DeBolt C, Lindquist S, et al. First Case of 2019 Novel Coronavirus in the United States. N Engl J Med 2020;382:929-36. [Crossref] [PubMed]
- Russell CD, Millar JE, Baillie JK. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet 2020;395:473-5. [Crossref] [PubMed]
- Shang L, Zhao J, Hu Y, et al. On the use of corticosteroids for 2019-nCoV pneumonia. Lancet 2020;395:683-4. [Crossref] [PubMed]
- Gautret P, Lagier JC, Parola P, et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents 2020.105949. [Crossref] [PubMed]
- Gautret P, Lagier JC, Parola P, et al. Clinical and microbiological effect of a combination of hydroxychloroquine and azithromycin in 80 COVID-19 patients with at least a six-day follow up: A pilot observational study. Travel Med Infect Dis 2020.101663. [Crossref] [PubMed]
- Molina JM, Delaugerre C, Le Goff J, et al. No evidence of rapid antiviral clearance or clinical benefit with the combination of hydroxychloroquine and azithromycin in patients with severe COVID-19 infection. Med Mal Infect 2020;50:384. [Crossref] [PubMed]
- Chen J, Liu L, Liu P, et al. A pilot study of hydroxychloroquine in treatment of patients with common coronavirus disease-19 (COVID-19). J Zhejiang Univ (Med Sci) 2020;49.
- Wang Y, Zhang D, Du G, et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet 2020;395:1569-78. [Crossref] [PubMed]
- Chen WH, Hotez PJ, Bottazzi ME. Potential for developing a SARS-CoV receptor-binding domain (RBD) recombinant protein as a heterologous human vaccine against coronavirus infectious disease (COVID)-19. Hum Vaccin Immunother 2020;16:1239-42. [Crossref] [PubMed]
- Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020;367:1260-3. [Crossref] [PubMed]
- Duan K, Liu B, Li C, et al. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc Natl Acad Sci U S A 2020;117:9490-6. [Crossref] [PubMed]
- Shen C, Wang Z, Zhao F, et al. Treatment of 5 Critically Ill Patients With COVID-19 With Convalescent Plasma. JAMA 2020;323:1582-9. [Crossref] [PubMed]
- Du L, He Y, Zhou Y, et al. The spike protein of SARS-CoV--a target for vaccine and therapeutic development. Nat Rev Microbiol 2009;7:226-36. [Crossref] [PubMed]
- WHO. Vaccine Development. 2020. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/global-research-on-novel-coronavirus-2019-ncov/solidarity-trial-accelerating-a-safe-and-effective-covid-19-vaccine


