Multifocal hepatocellular carcinoma: intrahepatic metastasis or multicentric carcinogenesis?
Hepatocellular carcinoma (HCC) is a frequent and deadly human disease, with 0.25-1 million new cases per year (1). HCC incidence varies with age, sex, and geographic region, the major number of cases coming from Asia, followed by Europe, Africa, North America, Latin America and Caribbean (2). The distribution of HCC cases among different populations reflects the differences in the exposition to different etiological factors. Hepatitis B virus (HBV) infection is a public health problem with approximately 2 billion people that have been exposed to the virus, and chronic HBV infection is the dominant risk factor for HCC in Eastern Asia and Sub-Saharan Africa (3). In recent years, a better knowledge of mutational and epigenetic events deregulating the signaling pathways involved in tumor progression, and the definition of genetic variants of HCC predicting disease outcome led to the identification of new potential prognostic markers and targets for molecular-based personalized therapies (4,5). Indeed, biomarkers with acceptable levels of sensitivity and specificity have been used, in recent years, to screen for and diagnose HCC, allowing in some cases early detection of HCC nodules in relatively high percentages of patients (6).
Multifocal hepatocarcinogenesis
Early HCC could be cured by surgical resection. However, a serious problem concerning this treatment is represented by the propensity to multifocal occurrence of this disease, which is responsible for frequent recurrences that largely influence its outcome. Multifocal HCC may be multiple HCCs of multicentric origin (MO), as well as intrahepatic metastases (IM) arising from a primary HCC (Figure 1). Different studies have documented that MO recurrences are more frequent than IM (7-9), but a recent report claims higher frequency and poorer prognosis of IM-type recurrences (10).
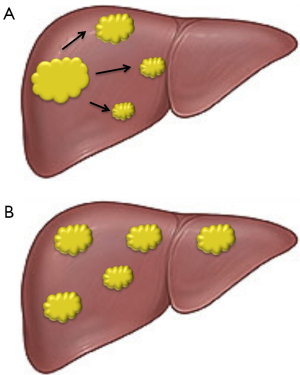
The differentiation of the two types of multifocal HCC is crucial because of their different clinical course and response to treatment. Numerous attempts to a differential diagnosis have been made in recent years. The comparison of clinicopathologic characteristics of MO and IM patients evidenced that MO HCC patients might have a favorable outcome compared to IM patients. Moreover, the presence of hepatitis B surface antigen (HBeAg), cumulative tumor size, tumor nodules location, cirrhosis, portal vein and/or microvascular tumor embolus and histological grade of the primary tumor may represent important discriminating factors (11). Other diagnostic criteria are based on the recognition of early recurrence, resulting from IM, and late recurrence indicating probable MO (12). The identification of patients at risk of recurrence is of prime importance to increase the chance of performing potentially curative interventions. Loss-of-heterozygosity analysis of specific DNA microsatellite loci has also been performed in the attempt to distinguish IM, of monoclonal origin, from multiclonal MO, probably arising from different preneoplastic lesions of cirrhotic liver (13,14) (Figure 1).
The omic approach
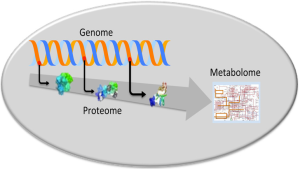
According to a common opinion the approaches merely based on morphologic and clinicopathologic criteria do not allow an accurate distinction between the different types of multifocal HCC (13,14). Therefore, new efforts have been recently dedicated to the research through omic analysis of new biologic mechanisms and molecular markers to better characterize multifocal HCC. The omic strategy includes the research of diagnostic and progression markers through genomic, proteomic and metabolomic analyses in order to evaluate gene, protein and metabolic deregulations (Figure 2).
The DNA microarray technology is currently used to identify specific gene-expression signatures of HCC. Recently this analysis has been exploited to predict early HCC recurrence due to intrahepatic metastasis (15). However, no well-defined predictors for late recurrence have been discovered (15). Some studies focused on the status of non-tumorous liver to predict late recurrence possibly due to de novo hepatocarcinogenesis, based on the idea of “field cancerization” (15), but not conclusive results have been obtained so far. In a recent study (16), the gene expression signature of metastatic primary HCCs has been found to be similar to that of their corresponding IMs, implying that genetic deregulation responsible for metastasis initiates in the primary tumors. Adrenomedullin (AM) gene has been identified, in the gene expression signature, as a leader gene overexpressed in HCCs with IM (16). A different gene expression profile was observed in multicentric HCCs (16).
One important new trend to delineate HCC biomarkers is the proteomic approach to identify proteins that differ in expression levels in liver tissue or in plasma during the progression from liver fibrosis, cirrhosis or steatohepatitis to HCC (17). The same approach has been used with reference to molecular diagnosis and metastatic recurrence of HCC (18). However, researches focusing on the study of the pathogenesis of IM and MO by proteomic approach are at their beginning. According to a recent report (19), a total of 1,025 and 900 spots indicative of protein overexpression have been found in expression profiles of patients with IM and MO, respectively. The spots indicative of decreased expression were 52 and 98, for IM and MO cases, respectively. The expression levels of 25 proteins were statistically different between the two groups of patients. Unfortunately this study has been done on a limited number of patients (10 IM and 5 MO cases) and the results have not been validated on a large patients’ cohort. Therefore, it is not possible to estimate if the newly identified proteins are actually potential biomarkers for identifying the multinodular HCC of clonal origin and discriminating between IM and MO cases.
A really innovative research, in this field, has been recently published by Miao and coworkers (20) who performed a multi-omics analysis to differentiate the MO vs. the IM disease, by decoding molecular differences between the two multifocal HCC models and recognize molecular markers for diagnosis and prognosis, as well as therapeutic targets. They evaluated the intrahepatic HCC lesions, and matched non-cancerous liver tissue and blood obtained from representative patients with HBV-related multifocal HCC who underwent tumor resection and exhibited distinct postsurgical courses. The samples were subjected to multi-omic analyses, integrating genomics and transcriptomics, and the results, further validated by a cohort of 174 HCC patients, were correlated with the clinicopathological data.
Two patients with multifocal HCC were identified. The patient I (PI) was cirrhotic and presented a multifocal poorly differentiated HCC. The patient II (PII) was non-cirrhotic and presented a well-differentiated multifocal HCC. It was hypothesized that PI was affected by an HCC with IM and PII by synchronous primary HCC development, with no spreading or metachronism. Tissues from multiple lesions for PI were: peripheral blood (PI-B), surrounding noncancerous liver (PI-N), primary HCC (PI-P), IM (PI-M1, PI-M2, and PI-M3), and a portal vein tumor thrombus (PI-V). Tissues from PII included: peripheral blood (PII-B), noncancerous liver (PII-N), and two HCCs located in the left (PII-L) and right lobes (PII-R). PI-N, PI-B, PI-P, PI-M1, PI-V, PII-N, PII-B, PII-L, and PII-R were used for next generation sequence, and the same tissues plus PI-M2 and PI-M3 were used for PCR validation. The determination of HBV integration in the different lesions clearly suggested the existence of different tumor clonality, in agreement with the different patterns of the multifocal tumors, in the two patients analyzed.
The evaluation of genomic alterations showed similar mutation patterns in all tumor tissues of PI, whereas the two HCCs of PII exhibited distinct mutation profiles. Further, significant enrichments of p53 signaling were present in all PI HCCs, whereas no cancer-related pathways were enriched in the PII tumors. Interestingly, the presence of the same deletions and amplifications in PI-P, PI-M1, and PI-V, with some differences between PI-M1 and PI-V, indicated the appearance of further selective mutations in HCC subclones during the formation of metastases. In PII HCCs different copy number variations occurred between PII-L and PII-R.
Importantly, the construction of a phylogenetic tree to predict the temporal development of each tissue, regardless of their germline differences, revealed that PI-M1-3 were phylogenetically most distant from the putative germline, with respect to PI-P, PI-V, or PI-N. Moreover, the sequence of PI HCCs development indicated that the portal vein tumor thrombus (PI-V) developed from the primary PI-I tumor and was followed by the metastatic lesions (PI-M1-3). These observations explained the genomic similarities of all PI HCCs indicating their origin from portal venous blood. Different patterns of the two PII HCCs were found, indicating that they were distant from germline and from each other, which implicates synchronous development of distinct clones.
These observations were in good agreement with the results of transcriptomic analysis that showed a stronger association of gene deregulation between the PI HCCs, indicative of genetic similarities between the metastases and the primary HCC in PI. In contrast, distinct patterns of transcriptomic dysregulation occurred in each PII HCCs, suggesting the existence of non-invasive phenotypes of PII HCCs developing from different premalignant clones of non-cirrhotic liver. Furthermore, it clearly appeared from the analysis of the deregulation of key genes and major signaling pathways, that neither PII HCC displayed molecular signatures of metastasis, which were instead found in PI HCCs. In complex, transcriptomic analysis substantiated the genetic alterations identified by genomic analysis.
Protein analysis also revealed the presence in all PI of similar metabolic alterations regarding changes in coenzyme metabolism and energy generation by mitochondrial oxidative phosphorylation, perturbations of carbohydrate catabolism and aerobic glycolysis, increases in nucleic acid metabolism, protein translation and transport, cell cycle, cell proliferation, and cell migration. Upregulation of genes involved in metastases formation, such as genes related to cytoskeletal remodeling and extracellular matrix organization was only found in PI-M1. In contrast, metabolic deregulation patterns were prevalently different in PII HCCs.
An important aspect of the study by Miao and co-workers (20) is the validation of the results of the multi-omic approach on a cohort of 174 patients with HBV-related HCC, in the attempt to identify new diagnostic and prognostic biomarkers for HCC. By the use of the BioCarta or KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways databases (21,22), the enrichment of pathways including cell cycle, p53 signaling, histidine metabolism, G2/M checkpoint, and Ran (Ras-related nuclear protein)-mediated mitotic spindle regulation was exclusively found in PI HCCs. Validation analysis confirmed the upregulation of six out of seven genes, such as: Histidine ammonia-lyase (HAL), stratifin, 14-3-3γ (SFN), kinesin superfamily protein 15 (KIF15), dual-specificity protein kinase (TTK), Budding uninhibited by benzimidazoles 1, s. cerevisiae, homolog of (BUB1), and minichromosome maintenance, s. cerevisiae, homolog of, 4 (MCM4). Different clinicopathologic features of HCCs were significantly associated with at least one of these genes. In addition, the evaluation of the relationships between the expression of above genes and the metastatic potential and postsurgical recurrence strongly suggested that TTK expression could be an independent prognostic indicator of HCC patients.
Conclusions and future perspectives
One of the important results of the multi-omic analysis of multifocal HCC, performed by Miao and coworkers (20) was the possibility to discern between the synchronous development of multicentric primary HCC and the metastatic disease. Furthermore, the integration of genomic and transcriptomic analyses with clinicopathologic features led to the identification of HCC biomarkers, validated with a large number of HCC patients.
In addition to the possibility to differentiate MO from IM, multi-omics profiling could provide essential information to evaluate the aggressiveness of existing lesions and apply personalized therapies, including postsurgical treatment. In this respect, it must be considered that the presence of several nodules in IM cases exhibiting various molecular alterations, indicative of differences in the progression of single lesions, requires multi-omic analysis of each new nodule in order to identify critical molecular lesions and to try their adjustment. Less aggressive MO cases may take advantage from the treatment of the underlying liver disease and from the resection of recurrences.
Another interesting result of the multi-omic approach performed by Miao and coworkers (20) is that the analysis of a single gene, TTK, led to the accurate prediction of early recurrence. This represents a valuable advantage with respect to gene-expression profiling studies that suggest multi-gene scores to predict recurrence and survival. In addition, when a specific genetic alteration predicting early recurrence is diffusely present in the liver, its detection by needle biopsies may allow chemopreventive strategies.
Although the multi-omics strategy potentially opens new groundbreaking perspectives for the diagnosis, prognosis and treatment of multi-focal HCC, it still presents some important limitations. The analysis regarded only HBV-HCC and should be extended to HCC caused by HCV infection, aflatoxin B1, alcoholism and metabolic diseases. Moreover, its clinical application could be seriously limited by the complexity of the analysis and the elevated costs. Nonetheless, the innovative results of the multi-omic approach propose new efforts to overcome these drawbacks.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- Llovet JM, Bruix J. Molecular targeted therapies in hepatocellular carcinoma. Hepatology 2008;48:1312-27. [PubMed]
- Ferlay J, Soerjomataram I, Ervik M, et al. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11. Lyon, France: International Agency for Research on Cancer; 2013. Available online: http://globocan.iarc.fr
- Mittal S, El-Serag HB. Epidemiology of hepatocellular carcinoma: consider the population. J Clin Gastroenterol 2013;47 Suppl:S2-6. [PubMed]
- Frau M, Feo F, Pascale RM. Pleiotropic effects of methionine adenosyltransferases deregulation as determinants of liver cancer progression and prognosis. J Hepatol 2013;59:830-41. [PubMed]
- Calvisi DF, Frau M, Tomasi ML, et al. Deregulation of signalling pathways in prognostic subtypes of hepatocellular carcinoma: novel insights from interspecies comparison. Biochim Biophys Acta 2012;1826:215-37.
- Han LL, Lv Y, Guo H, et al. Implications of biomarkers in human hepatocellular carcinoma pathogenesis and therapy. World J Gastroenterol 2014;20:10249-61. [PubMed]
- Chen YJ, Yeh SH, Chen JT, et al. Chromosomal changes and clonality relationship between primary and recurrent hepatocellular carcinoma. Gastroenterology 2000;119:431-40. [PubMed]
- Ochiai T, Urata Y, Yamano T, et al. Clonal expansion in evolution of chronic hepatitis to hepatocellular carcinoma as seen at an X-chromosome locus. Hepatology 2000;31:615-21. [PubMed]
- Nomoto S, Kinoshita T, Kato K, et al. Hypermethylation of multiple genes as clonal markers in multicentric hepatocellular carcinoma. Br J Cancer 2007;97:1260-5. [PubMed]
- Wang B, Xia CY, Lau WY, et al. Determination of clonal origin of recurrent hepatocellular carcinoma for personalized therapy and outcomes evaluation: a new strategy for hepatic surgery. J Am Coll Surg 2013;217:1054-62. [PubMed]
- Li SL, Su M, Peng T, et al. Clinicopathologic characteristics and prognoses for multicentric occurrence and intrahepatic metastasis in synchronous multinodular hepatocellular carcinoma patients. Asian Pac J Cancer Prev 2013;14:217-23. [PubMed]
- Du ZG, Wei YG, Chen KF, et al. Risk factors associated with early and late recurrence after curative resection of hepatocellular carcinoma: a single institution's experience with 398 consecutive patients. Hepatobiliary Pancreat Dis Int 2014;13:153-61. [PubMed]
- Morimoto O, Nagano H, Sakon M, et al. Diagnosis of intrahepatic metastasis and multicentric carcinogenesis by microsatellite loss of heterozygosity in patients with multiple and recurrent hepatocellular carcinomas. J Hepatol 2003;39:215-21. [PubMed]
- Ng IO, Guan XY, Poon RT, et al. Determination of the molecular relationship between multiple tumour nodules in hepatocellular carcinoma differentiates multicentric origin from intrahepatic metastasis. J Pathol 2003;199:345-53. [PubMed]
- Utsunomiya T, Shimada M, Imura S, et al. Molecular signatures of noncancerous liver tissue can predict the risk for late recurrence of hepatocellular carcinoma. J Gastroenterol 2010;45:146-52. [PubMed]
- Nakata T, Seki N, Miwa S, et al. Identification of genes associated with multiple nodules in hepatocellular carcinoma using cDNA microarray: multicentric occurrence or intrahepatic metastasis? Hepatogastroenterology 2008;55:865-72. [PubMed]
- Beretta L. Comparative analysis of the liver and plasma proteomes as a novel and powerful strategy for hepatocellular carcinoma biomarker discovery. Cancer Lett 2009;286:134-9. [PubMed]
- Qin LX, Tang ZY. Recent progress in predictive biomarkers for metastatic recurrence of human hepatocellular carcinoma: a review of the literature. J Cancer Res Clin Oncol 2004;130:497-513. [PubMed]
- Su M, Li LQ, Peng T, et al. Comparative proteomic approach in differentiating multicentric occurrence and intrahepatic metastasis in multinodular hepatocellular carcinomas. Chin J Cancer 2010;29:52-8. [PubMed]
- Miao R, Luo H, Zhou H, et al. Identification of prognostic biomarkers in hepatitis B virus-related hepatocellular carcinoma and stratification by integrative multi-omics analysis. J Hepatol 2014;61:840-9. [PubMed]
- Merico D, Isserlin R, Stueker O, et al. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 2010;5:e13984. [PubMed]
- Nishimura D. BioCarta. Biotech Software Internet Rep 2001;2:117-20.


