
The Ras-ERK signaling pathway regulates acetylated activating transcription factor 2 via p300 in pancreatic cancer cells
Introduction
Pancreatic cancer is among the most common cancers and presents a major challenge for clinicians due to its asymptomatic nature. Typically, diagnosis is only possible in advanced stages of the disease and this reduces survival rates considerably. Almost 95% of pancreatic cancers are exocrine in nature, and among these, the most common is pancreatic ductal adenocarcinoma (PDAC) (1-3). As for treatment, surgery is the only potentially curative treatment for pancreatic adenocarcinoma; unfortunately, at most only 20% of newly diagnosed patients have disease suitable for curative resection after careful pretherapeutic staging (4). After curative resection for pancreatic cancer, 69–75% of patients relapse within 2 years and 80–90% within 5 years (5,6). This high rate of disease recurrence following surgery provides a strong rationale for the need for adjuvant therapy and chemoradiotherapy (CRT) to improve overall survival following resection (7). Gemcitabine, a standard care cytotoxic nucleoside analog, is used very scarcely and achieves a median survival time of only 5 months (8,9). Treatment with a combination of chemotherapeutic agents and targeting of specific molecular pathways can significantly improve the outcomes. In this regard, the extracellular signal-regulated protein kinases (ERK) 1 and 2 are currently being explored (9,10).
The Ras-ERK signaling pathway, which controls cell proliferation, survival, differentiation, and motility, is often found to be upregulated in pancreatic cancer (11). Although the mechanism of epigenetic regulation is not fully understood, it is known that the mutations or growth factors activate the three-tiered Ras-ERK signaling pathway. In this case, Ras guanosine triphosphatase (GTPase) acts upstream of MEK (mitogen-activated protein kinase), which is upstream of ERK (12-15). In the ductal adenocarcinoma model, inhibition of the Ras-ERK pathway blocks cellular proliferation and limits metastasis. However, in the development of novel therapeutics, targeting of downstream kinases is preferable to oncogenic RAS (16).
Activating transcription factor-2 [ATF2, also known as cAMP response element-binding protein 1 (CRE-BP1)] is a helix-loop-helix transcription factor that has been associated with a various cancers due to its role in maintaining the cancer cell phenotype (17-20). Physically distinct, functional domains of ATF2 impart sequence-specific deoxyribonucleic acid (DNA) binding and regulate transcriptional functions (21). The Ras signal cascade phosphorylates ATF2 to promote carcinogenesis by altering the transcription of downstream genes. However, a dominant-negative ATF2 could block cancer cell proliferation (22). P300 is one of global transcriptional coactivators that turns tightly wrapped around a central histone octamer is involved in the regulation of various DNA-binding transcriptional factors (23). P300 and CREB-binding protein (CBP), as global transcriptional coactivators are capable of histone acetyltransferase activity (24). It is known that p300 performs acetylation of ATF2 at the Lys357 position (25), which is important for the downstream histone modification at the target binding sites. However, its role in pancreatic cancer is still unknown.
In this study, we report that the Ras-ERK signaling pathway regulates acetylated ATF2 (ATF2K357ac) by inducing proteasome-dependent degradation of the p300 protein. Interestingly, we found that p300 was degraded only at the protein level without affecting its transcription status. This in turn affects the expression of downstream genes that are involved in maintaining the pancreatic cancer cell phenotype. Overall, our results suggest a potential mechanism for the regulation of ATF2K357ac in the progression of pancreatic cancer.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-5880).
Methods
Cells culture and transfection
ASPC-1 and BXPC-3 pancreatic cancer cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, USA). The cells were maintained at 37 °C with 5% carbon dioxide (CO2) in Dulbecco’s Modified Eagle Medium (DMEM) with 10% fetal bovine serum (FBS), 100 µg/mL streptomycin, and 100 units/mL penicillin. These cells were grown overnight to achieve transfection of the respective plasmids and small interfering ribonucleic acids (siRNAs) using lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. The cells were utilized for the experiments after 48 h of transfection.
Plasmids and siRNA
Plasmids were generated for the respective coding regions of human ATF2, p300, histone deacetylase 2 (HDAC2), mouse double minute 2 homolog (MDM2), and Ras [wild-type Kirsten (K)] from the pancreatic cancer cells (ASPC-1 and BXPC-3) using polymerase chain reaction (PCR) amplification. All plasmids were confirmed by sequencing. Mutant enhanced green fluorescent protein (pEGFP)-RasG12V was obtained using site-directed mutagenesis, and the resultant template (pEGFP-RasG12V) was used to construct the double mutant pEGFP-RasG12V/T35S. HDAC2 (Shanghai GenePharma), ATF2, and ATF2K357Q were also generated (TaKaRa Mutant BEST Kit).
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
For ribonucleic acid (RNA) isolation, cell lysis was performed in the tissue culture plates using TRIzol reagent (Invitrogen), and subsequently subjected to deoxyribonuclease I (DNase-I) treatment. Using the respective oligo-deoxythymine (dT) primers (Invitrogen) and Moloney Murine Leukemia Virus (M-MuLV) reverse transcriptase (Fermentas), synthesis of the first-strand complementary DNA (cDNA) was conducted from the total RNA. Real-time PCR was performed using a QuantiTect SYBR Green PCR Kit (Qiagen). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control. All samples were analyzed in triplicate according to the manufacturer’s instructions. The respective primer sequences are provided in the supplementary data (Table S1).

Full table
Western blot analysis
The cells were washed in phosphate-buffered saline (PBS) and lysis was carried out in the radioimmunoprecipitation assay (RIPA) buffer on ice. Lysates were centrifuged to obtain the supernatants and the total protein concentrations were determined. These were then transferred to the nitrocellulose membranes, followed by incubation with anti-ATF2 antibody (1:500), anti-ERK1/2 antibody (1:500), anti-phospho-ERK1/2T202 antibody (1:500), anti-HDAC2 antibody (1:500), anti-hemagglutinin (HA) antibody (1:500), anti-green fluorescent protein (GFP) antibody (1:500), anti-histone (His) H3 antibody (1:500), anti-p300 antibody (1:500), anti-MDM2 antibody (1:500), and anti-β-actin antibody (1:1,000) at 4oC overnight. All antibodies were purchased from Abcam (Abcam, Cambridge, UK). After this, incubation with secondary horseradish peroxidase (HRP)-conjugated anti-mouse antibodies was conducted for 1 h. Protein bands were illuminated using the luminal agent SC-2048 (Santa Cruz Biotechnology, California, USA) according to the manufacturer’s instructions.
Cell viability [cell counting Kit-8 (CCK-8)] assay
The cells were seeded and incubated for 24, 48, 72, and 96 h in 96-well culture plates. These were mixed and incubated with CCK-8 (Thermo Fisher Scientific, USA) for another 3.5 h to assess the transfected cells (5×103 cells/well). The absorbance of each well was measured at 450 nm using an Emax spectrophotometer (Thermo Fisher Scientific).
Cell proliferation (colony formation) assay
For the soft-agar colony formation assay, the transfected cells were suspended in DMEM containing 0.35% low-melting agarose. These were then plated onto solidified 0.6% agarose in DMEM in six-well culture plates at a density of 1×103 cells per well. After 3 weeks of incubation, the number of colonies was observed microscopically.
Transwell (cell migration) assay
The cell migration was measured using the transwell migration assay. 200 µL suspensions of transfected cells (1×104 cells/well) in serum-free medium were added to the upper chambers and 600 µL DMEM containing 10% FBS was added into the lower chambers. After 48 h of incubation, migrated cells were fixed with methanol and stained with crystal violet. Finally, the stained cells were counted using a light microscope (Olympus Corporation, Tokyo, Japan).
Flow cytometry and cell apoptosis
Cell apoptosis was detected using the Annexin V-FITC kit (Biosea Biotechnology Co., Beijing, China). The transfected cells, resuspended in PBS buffer (5.0×105 cells/mL), were double-stained with Annexin V-Alexa Fluor 647 and propidium iodide (PI). The apoptotic rate was measured using a flow cytometer (BD Biosciences, USA).
Chromatin immunoprecipitation assay (ChIP)
Sample (3×106 transfected cells/sample) cross-linking was carried out in 1% formaldehyde for 10 min at room temperature (RT). Samples were washed twice with ice cold PBS prior to lysis in sodium dodecyl sulfate (SDS) Lysis Buffer (Upstate, # 20–163). Lysates were then sonicated (Ultrasonic bath, Bioruptor, Diagenode) to shear the DNA to an average length of 200–800 base pairs (bp). These were then centrifuged at 15000 g to collect the supernatants, followed by dilution in ChIP Dilution Buffer (Upstate, #20–153). Immunoprecipitation was carried out overnight with 2 µg rabbit anti-ATF2K357ac antibody (synthesized by Cell Signaling Technology, Danvers, Massachusetts, USA) at 4 °C. For the control, immunoprecipitations were performed using 2 µg normal anti-immuoglobulin G (IgG) antibody. The beads were subjected to three sequential washes (5 min each at 4 °C) in the following buffers: low-salt (Upstate, #20–154), high-salt (Upstate, #20–155), and lithium chloride (LiCl) (Upstate, #20–156). Lastly, two further washing steps were performed with Tris- Ethylenediaminetetraacetic acid (TE) (1×; Upstate, #20–157) for 2 min at RT, and DNA was eluted in 1% SDS/100 mM sodium bicarbonate (NaHCO3) for 15 min at RT. Crosslinking was reversed in 200 mM sodium chloride (NaCl) for 7 h at 65 °C. The eluted DNA was precipitated with ethanol at −20 °C overnight. Samples were analyzed by quantitative real-time polymerase chain reaction (qRT-PCR) using 1.5 µL immunoprecipitated DNA and the serial dilutions of 10% input DNA (1:4, 1:20, 1:100, and 1:500).
Statistical analysis
Data analysis was carried out using SPSS 20.2 (SPSS, Chicago, IL, USA) and GraphPad Prism 7 (La Jolla, CA, USA) software, and Student’s (two-tailed) t-test or a one-way analysis of variance (ANOVA) was applied. Data were presented as mean ± standard deviation (SD). A minimum of three experiments were completed independently. P<0.05 was considered statistically significant.
Results
Acetylated ATF2 is regulated by the Ras-ERK pathway
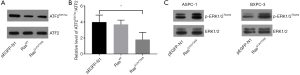
Western blot analysis of the transfected cells showed that compared to the control (empty vector, pEGFP-N1) or Ras-wild-type (WT) (pEGFP-RasWT), transfection of the double mutant K-RasG12V/T35S (pEGFP-RasG12V/T35S) significantly downregulated the level of acetylated ATF2 protein (ATF2K357ac) (P<0.05) (Figure 1A,B). Also, when the activation of mitogen-activated protein kinase (MAPK) was measured by detecting the level of phosphorylated ERK1/2 (p-ERK1/2Thr202), we found that activation was only evident in the RasG12V/T35S-expressing cells, indicating the activation of the downstream pathway. Here, total ERK1/2 proteins were used as loading controls (Figure 1C). Overall, these results indicate that acetylated ATF2 is regulated by the Ras-ERK pathway.
Mutant ATF2K357Q inhibits cell proliferation and migration mediated by Ras-ERK pathway
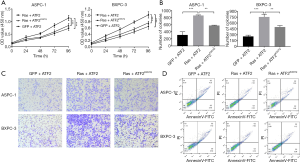
A lysine-to-glutamine (K→Q) substitution is a known acetyl mimic that has been used to study the effect of acetyl modification in many acetyltransferases. It is a stable modification that inhibits deacetylation. In this study, we created an ATF2K357Q mutant to mimic the acetylated form of ATF2. To measure the effects of acetylated ATF2 on cell viability, colony formation, migration, and apoptosis, we used CCK-8, soft-agar, transwell migration, and flow cytometry assays, respectively. The results from these assays demonstrated that expression of Ras and ATF2 increased cell viability (Figure 2A), cell numbers (Figure 2B), and cell migration (Figure 2C). Although, cell apotosis decreased (P<0.01) (Figure 2D) in the ASPC-1 and BXPC cells. However, the transfection of ATF2K357Q (acetylated, inactive ATF2) in place of ATF2 restored the cell phenotype (Figure 2A,B,C,D). An empty vector (GFP) along with ATF2 was used as a control. These results strongly indicate that the acetylation of ATF2 is regulated by the Ras-ERK pathway in pancreatic cancer cells.
Acetylation of ATF2 regulates transcription of the downstream gene of the Ras-ERK signaling pathway
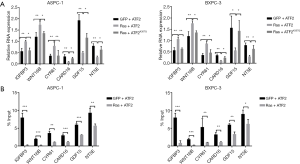
Genes (IGFBP3, WNT16B, CYR61, CARD16, GDF15, and NT5E) downstream to the Ras-ERK pathway are upregulated in cancer. To investigate the transcriptional effect of ATF2 acetylation on these genes, qRT-PCR studies were conducted. The results demonstrated that the co-transfection of ATF2K357Q along with Ras decreased transcription of the target genes compared to ATF2 (Figure 3A). Furthermore, in ChIP assays, we observed that ATF2 was reduced on the promoters of respective genes (Figure 3B). These results imply that ATF2 binding to the promoter region of the aforementioned genes is required for transcriptional regulation.
The silencing of HDAC2 attenuates acetylated ATF2-dependent phenotypes of pancreatic cancer cells
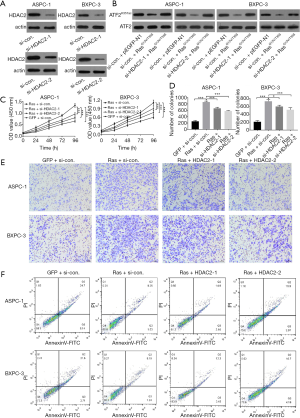
HDAC families of deacetylases are capable of removing acetylation modification. We aimed to explore whether HDAC2 silencing would increase the level of acetylated ATF2. For this, two siRNAs (si-HDAC2-1 and si-HDAC2-2) were synthesized to silence HDAC2, and si-con siRNA was used as a negative control. As expected, PCR results showed that the HDAC2 messenger RNA (mRNA) level was remarkably reduced only in si-HDAC2-1 or si-HDAC2-2 transfected cells but not in the case of si-con siRNA (Figure 4A). Furthermore, using western blot analysis, we examined the effect of HDAC2 silencing on the levels of acetylated ATF2 upon activation of the Ras-ERK pathway. We found that HDAC2 silencing decreased the levels of ATF2K357ac in both types of pancreatic cancer cells (Figure 4B). In addition, the CCK-8, colony formation, and transwell migrations assays also demonstrated that the cell viability, proliferation, and migration enhanced by the Ras-ERK pathway were notably repressed upon silencing of HDAC2 (P<0.001) (Figures 4C,D,E), whereas apoptosis was elevated (Figure 4F). Altogether, these results emphasize the importance of acetylated ATF2 in maintaining cancer cell phenotypes.
ATF2K357ac is downregulated by p300 degradation, induced by the Ras-ERK pathway
The p300 protein is a coactivator that has intrinsic acetyltransferase activity and is known to interact with ATF2. Western blot analysis was performed to investigate whether p300 was involved in regulating the levels of ATF2K357ac. Our results showed that HDAC2 and p300 protein levels were reduced upon activation of the Ras-ERK pathway (Figures 5A,B). The reduced level of p300 downregulated the ATF2K357ac in the ASPC-1 and BXPC-3 cells (Figure 5C). However, we found that the mRNA level of p300 and HDAC2 was not affected (Figure 5D). Subsequently, ChIP assays revealed that p300 could bind to the promoter region of downstream target genes (CAFRD16, CYR61, WNT16B, IGFBP3, GDF15, and NT5E) regulated by ATF2, however was downregulated after pathway activation, along with the downregulation of ATF2 (Figure 5E).
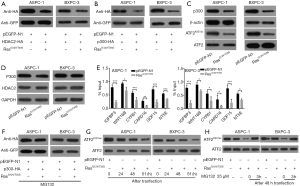
To further confirm the involvement of p300 in regulation, MG132 proteasome inhibitor was added into ASPC-1 and BXPC-3 cells. In contrast to Figure 5B, p300 levels were not decreased (Figure 5F). Upon activation of the Ras-EKR pathway (using RasG12V/T35S in the absence of MG132), we noticed a reduction in the levels of ATF2K357ac at 48 h post-transfection (Figure 5G). However, after 3 h of MG132 addition, the level of ATF2K357ac recovered (Figure 5H). These results indicated that the degradation of p300 downregulated ATF2K357ac.
p300 degradation was mediated by MDM2, induced by the Ras-ERK pathway
E3 ubiquitin ligase MDM2 is associated with the degradation of various histone acetylates. So, we investigated the possible involvement of MDM2 in the degradation of p300 following activation of the Ras-ERK pathway. ASPC-1 and BXPC-3 cells were transfected with MDM2-His along with K-RasG12V/T35S. After 48 h of transfection, the levels of exogenous and endogenous p300 were detected using anti-HA and anti-p300 antibodies, respectively. The results showed that RasG12V/T35S and MDM2 could significantly reduce p300 protein levels (Figures 6A,B). MDM2C464A, a really interesting new gene (RING)-finger domain mutant lacks ubiquitin activity. So, when MDM2C464A (MDM2-MU) was transfected, protein levels of p300 did not change significantly (Figures 6C,D), indicating that the degradation of p300 was dependent on MDM2.
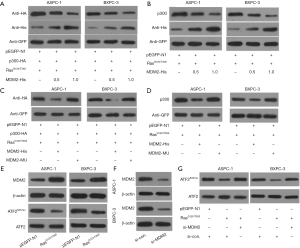
Furthermore, we analyzed whether activation of the Ras-ERK pathway upregulated MDM2 to enhance p300 degradation. Results showed that activation of the Ras-ERK pathway significantly increased the level of endogenous MDM2 (Figure 6E). To examine whether the upregulation of MDM2 increased p300 degradation, the MDM2 level was decreased by using siRNA (si-MDM2) against MDM2 (Figure 6F). Interestingly, the downregulation of ATF2K357ac was inhibited after the transfection of si-MDM2 (Figure 6G). Overall, these results suggested that the Ras-ERK pathway, which regulates ATF2K357ac induced p300 degradation through MDM2.
Discussion
A hyper-activated Ras-ERK pathway is closely associated with pancreatic cancer, particularly in the regulation of cell growth and migration (9,26). In this study, Ras was found to be an important upstream signaling molecule that upregulates certain genes downstream of the Ras-ERK pathway. These genes, including CYR61, WNT16B, IGFBP3, GDF15, NT5E, and CARD16, are regulators of cancer cell proliferation and migration activity (27).
ATF2 is also an epigenetic regulator that possesses intrinsic acetyltransferase activity. Specifically, it acts to acetylate the histones H2B and H4. The Ras-ERK pathway-mediated phosphorylation of ATF2 not only triggers its DNA-binding effectiveness but also augments its intrinsic acetyltransferase activity (21). The regulatory effect of ATF2 on histone acetylation can be extended to many amino acid-regulated genes (28). Interestingly, ATF2 can also be acetylated by another histone acetyltransferase, p300, at the two lysine acetylation sites, K357 and K374. These lysine residues are located in the basic leucine zipper (b-ZIP) domain of ATF2. The modification at this site(s) triggers conformational changes to alter the DNA-binding capacity of ATF2. Importantly, mutations of the b-ZIP acetylation site hinder transcriptional activation of ATF2 (25).
Numerous cancer studies have found that ATF2 is an important regulator in pancreatic cancer. However, the specific role of ATF2K357ac is unclear. In this study, we demonstrated that the ATF2K357ac regulates the characteristic phenotype of pancreatic cancer cells via the Ras-ERK pathway by regulating the expression of downstream proliferation, migration, and other related genes. We found that the level of ATF2K357ac is strongly associated with the development of pancreatic cancer.
It is reported that HDAC2, a deacetylase, can reverse-regulate ATF2 (25). Hence, we investigated the effect of HDAC2 on the proliferation and migration of pancreatic cells using siRNA experiments, and found that the cell phenotypes are related to the expression level of HDAC2, and HDAC2 regulates ATF2K357ac levels. Furthermore, the activated Ras-ERK pathway induces the degradation of p300 mediated by the MDM2-dependent proteasome pathway, which in turn regulates the level of ATF2K357ac.
Conclusions
However, there are some limitations to this regulatory model of ATF2 due to the presence of other enzyme molecules, especially deacetylases. These may also simultaneously participate in regulating ATF2K357ac by altering the acetyl modification site. Moreover, the affinity of p300 and HDAC2 to ATF2 is unknown, especially in the Ras-ERK signaling pathway-mediated regulation. The differential affinities of p300 or HDAC2 for ATF2 could be significant. It is also necessary to investigate whether the localization of p300 is altered, as it could have downregulated ATF2K357ac due to the decreased availability of p300, thereby p300 could change the affinity for ATF2. Thus, these aspects need to be explored further to comprehensively understand the regulation of ATF2K357ac.
Acknowledgments
Funding: This study was supported by the Natural Science Foundation of Liaoning Province [No. 180530068], the Liaoning Province Project of Hundred, Thousand, Ten Thousand Talent Projects [3200417003], and the Project of Shenyang Municipal Science and Technology Bureau [17231178].
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-5880
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-5880
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-5880). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Shi J, Xue J. Inflammation and development of pancreatic ductal adenocarcinoma. Chin Clin Oncol 2019;8:19. [Crossref] [PubMed]
- Haeberle L, Esposito I. Pathology of pancreatic cancer. Transl Gastroenterol Hepatol 2019;4:50. [Crossref] [PubMed]
- Feng J, Rao M, Wang M, et al. Triptolide suppresses pancreatic cancer cell proliferation by inhibiting hedgehog signaling pathway activity. Sci China Life Sci 2019;62:1409-12. [Crossref] [PubMed]
- Ducreux M, Cuhna AS, Caramella C, et al. Cancer of the pancreas: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Annals of oncology: official journal of the European Society for Medical Oncology. 2015;26 Suppl 5:v56-68. [Crossref] [PubMed]
- Pointer DT Jr, Al-Qurayshi Z, Hamner JB, et al. Factors leading to pancreatic resection in patients with pancreatic cancer: a national perspective. Gland Surg 2018;7:207-15. [Crossref] [PubMed]
- Sinn M, Bahra M, Liersch T, et al. CONKO-005: Adjuvant Chemotherapy With Gemcitabine Plus Erlotinib Versus Gemcitabine Alone in Patients After R0 Resection of Pancreatic Cancer: A Multicenter Randomized Phase III Trial. J Clin Oncol 2017;35:3330-7. [Crossref] [PubMed]
- Conroy T, Ducreux M. Adjuvant treatment of pancreatic cancer. Curr Opin Oncol 2019;31:346-53. [Crossref] [PubMed]
- O'Reilly EM, Abou-Alfa GK. Cytotoxic therapy for advanced pancreatic adenocarcinoma. Semin Oncol 2007;34:347-53. [Crossref] [PubMed]
- Zheng C, Jiao X, Jiang Y, et al. ERK1/2 activity contributes to gemcitabine resistance in pancreatic cancer cells. J Int Med Res 2013;41:300-6. [Crossref] [PubMed]
- Kohno M, Pouyssegur J. Targeting the ERK signaling pathway in cancer therapy. Ann Med 2006;38:200-11. [Crossref] [PubMed]
- Chadha KS, Khoury T, Yu J, et al. Activated Akt and Erk expression and survival after surgery in pancreatic carcinoma. Ann Surg Oncol 2006;13:933-9. [Crossref] [PubMed]
- Cao AL, Tang QF, Zhou WC, et al. Ras/ERK signaling pathway is involved in curcumin-induced cell cycle arrest and apoptosis in human gastric carcinoma AGS cells. J Asian Nat Prod Res 2015;17:56-63. [Crossref] [PubMed]
- Dorard C, Vucak G, Baccarini M. Deciphering the RAS/ERK pathway in vivo. Biochem Soc Trans 2017;45:27-36. [Crossref] [PubMed]
- Santos E, Crespo P. The RAS-ERK pathway: A route for couples. Sci Signal 2018;11:eaav0917. [Crossref] [PubMed]
- Neuzillet C, Tijeras-Raballand A, de Mestier L, et al. MEK in cancer and cancer therapy. Pharmacol Ther 2014;141:160-71. [Crossref] [PubMed]
- Neuzillet C, Hammel P, Tijeras-Raballand A, et al. Targeting the Ras-ERK pathway in pancreatic adenocarcinoma. Cancer Metastasis Rev 2013;32:147-62. [Crossref] [PubMed]
- Wu DS, Chen C, Wu ZJ, et al. ATF2 predicts poor prognosis and promotes malignant phenotypes in renal cell carcinoma. J Exp Clin Cancer Res 2016;35:108. [Crossref] [PubMed]
- Zhang R, Luo H, Wang S, et al. MiR-622 suppresses proliferation, invasion and migration by directly targeting activating transcription factor 2 in glioma cells. J Neurooncol 2015;121:63-72. [Crossref] [PubMed]
- Lo Iacono M, Monica V, Vavalà T, et al. ATF2 contributes to cisplatin resistance in non-small cell lung cancer and celastrol induces cisplatin resensitization through inhibition of JNK/ATF2 pathway. Int J Cancer 2015;136:2598-609. [Crossref] [PubMed]
- Zhang S, Dong X, Ji T, et al. Long non-coding RNA UCA1 promotes cell progression by acting as a competing endogenous RNA of ATF2 in prostate cancer. Am J Transl Res 2017;9:366-75. [PubMed]
- Kawasaki H, Schiltz L, Chiu R, et al. ATF-2 has intrinsic histone acetyltransferase activity which is modulated by phosphorylation. Nature 2000;405:195-200. [Crossref] [PubMed]
- Vlahopoulos SA, Logotheti S, Mikas D, et al. The role of ATF-2 in oncogenesis. Bioessays 2008;30:314-27. [Crossref] [PubMed]
- Janknecht R, Hunter T. Transcription. A growing coactivator network. Nature 1996;383:22-3. [Crossref] [PubMed]
- Ogryzko VV, Schiltz RL, Russanova V, et al. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 1996;87:953-9. [Crossref] [PubMed]
- Karanam B, Wang L, Wang D, et al. Multiple roles for acetylation in the interaction of p300 HAT with ATF-2. Biochemistry 2007;46:8207-16. [Crossref] [PubMed]
- Liu Y, Wang DL, Chen S, et al. Oncogene Ras/phosphatidylinositol 3-kinase signaling targets histone H3 acetylation at lysine 56. J Biol Chem 2012;287:41469-80. [Crossref] [PubMed]
- Li Y, Sun W, Sun D, et al. Ras-ERK1/2 signaling promotes the development of uveal melanoma by downregulating H3K14ac. J Cell Physiol. 2019. [Epub ahead of print]. [Crossref] [PubMed]
- Bruhat A, Chérasse Y, Maurin AC, et al. ATF2 is required for amino acid-regulated transcription by orchestrating specific histone acetylation. Nucleic Acids Res 2007;35:1312-21. [Crossref] [PubMed]


