
Correlation between the coexpression of zinc finger and SCAN domain-containing protein 31 and transcriptional activator with PDZ-binding motif and prognosis in hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) ranks as the third leading cause of cancer-related mortality worldwide and as the second-placed cause in China (1,2). However, HCC treatment has some limitations, resulting in a high rate of recurrence after liver resection, as well as a poor prognosis. Therefore, to improve the prognosis of HCC patients, new therapeutic targets desperately need to be discovered.
As one of the nuclear effectors of Hippo-related signaling pathways, transcriptional coactivator with PDZ binding motif (TAZ), which is also referred to as WW-domain-containing transcriptional regulator protein 1 (WWTR1) (3), regulates the control of organ size, cancer stem cell properties, and epithelial-mesenchymal transition (EMT) (4-8). In our previous study, TAZ was found to be related to the survival, proliferation, and EMT of HCC cells, as well as survival, apoptosis, and drug resistance in intrahepatic cholangiocarcinoma (ICC) cells (9,10).
In the present study, we analyzed the potential target genes of TAZ after its knockdown. Among the messenger RNA (mRNA) profiles, the expression of ZSCAN31 was observed to be decreased when TAZ was down-regulated. However, few studies to date have focused on the role of ZSCAN31 in tumorigenesis, especially in HCC. In this study, we also evaluated the change in ZSCAN31 expression and determined its effect on survival in HCC patients treated with surgical resection.
The results showed that ZSCAN31 expression was significantly higher in HCC tissues, which indicated a poor prognosis. The combination of TAZ/ZSCAN31 could serve as a novel predictor of prognosis in HCC as well as a potential target for HCC therapy after resection. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-6373).
Methods
Study subjects
Clinical tissue samples were collected from 83 HCC patients (70 males and 13 females) who underwent liver resection in the First Affiliated Hospital of Chongqing Medical University between Jan, 2006 and Dec, 2010. Signed informed consent forms were obtained from all patients. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Clinical Specimens Ethics Committee of the First Affiliated Hospital of Chongqing Medical University.
Genome-wide gene expression array
HCC cells (HepG2) were treated with si-NC or si-TAZ for 2 days. The gene expression profiles were detected using an Affymetrix Human U133 Plus 2.0 Array (Shanghai Biotechnology Corporation) in accordance with the manufacturer’s instructions.
The Cancer Genome Atlas (TCGA) database
Fifty-one normal liver tissues and 270 HCC tissues were obtained from the TCGA database (https://tcga-data.nci.nih.gov/tcga/) to validate mRNA expression after TAZ knockdown. A corresponding heat map of different mRNA expression was constructed using the heatmap package in R language. The prognostic significance of the mRNAs for overall survival (OS) was also evaluated, using Cox regression analysis and the Kaplan-Meier method, with the log-rank test used to compare the survival curves The corresponding clinical data of 179 HCC cases were downloaded from the Cancer Browser (https://genome-cancer.ucsc.edu/). This study was carried out in accordance with the publication guidelines of TCGA.
Immunohistochemistry
Immunohistochemistry (IHC) was carried out according to the procedures and regents detailed in our previous study (11). Tissue sections were incubated with human antibodies for TAZ and ZSCAN31 (dilution 1:500) at 4 °C for 24 hours. Then, the sections were cultivated by adding horseradish peroxidase (HRP) at room temperature for 20–30 min, with 3,3’-Diaminobenzidine (DAB) as the color development reagent and hematoxylin for counterstaining. Immunoreactive density of ZSCAN31 was detected using Imagepro Plus 6.0 software.
Co-immunoprecipitation (Co-IP)
Co-immunoprecipitation was performed as previously described (12). The experiment was carried out using a Dynabeads® Co-immunoprecipitation Kit in strict accordance with the user guide. Protein was extracted by resuspending cells in extraction buffer. The cell supernatant was then collected. For each antibody-coupled bead, beads were resuspended in cell lysate at 2–8 °C for 20–30 min. After that, the beads were rinsed with extraction buffer and last wash buffer, before being resuspended in elution buffer and incubated for 5 min. The elute liquid contained the purified protein complex. Western blot was performed to detect the proteins.
Statistical analysis
Statistical analyses were carried out using SPSS 17.0. The potential correlation between TAZ/ZSCAN31 coexpression and HCC clinicopathological parameters was examined by chi-square test or Fisher’s exact test. The Kaplan-Meier method was used to measure OS and disease-free survival, and the survival curves were compared with the log-rank test. P<0.05 indicated a statistically significant difference.
Results
Potential target genes of TAZ
The potential target genes of TAZ were analyzed with a genome-wide gene expression array. All altered genes were detected in TCGA database. The shared genes among following three groups were identified to further analyze the potential target genes of TAZ: (I) significantly changed genes in HCC cells after TAZ knockdown (274 genes; Figure 1A); (II) differentially expressed genes in TCGA database (67 genes; Figure 1B); and (III) genes in TCGA database predicted to have a prognostic value (20 genes). Furthermore, ZSCAN31 was highly expressed in HCC tissues and was revealed to be a poor prognostic factor in HCC after surgical resection (Figure 1B,C). As shown in Figure 2A, two genes were shared by these three groups. ZSCAN31 and DUSP1 were observed to be down-regulated in the HCC cells after TAZ knockdown (Figure 2B), which was consistent with our previous study’s findings on the function of TAZ in HCC. However, the TCGA analysis in our previous study revealed dual specificity phosphatase 1 (DUSP1) to have the opposite tendency to TAZ (9). Therefore, in this study, ZSCAN31 was chosen as our terminal target.
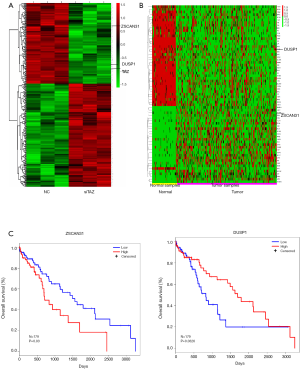

A significant change in ZSCAN31 mRNA expression was identified (fold change =2.24 and P=0.037) after TAZ knockdown (Figure 2B). In TCGA database, ZSCAN31 expression was significantly increased (fold change =2.52 and P<0.001) in HCC tissues, indicating a poor prognosis (HR =1.8 and P=0.032) after surgical resection for HCC.
High ZSCAN31 expression was associated with low OS after surgical resection in HCC
To verify the results of the gene array, reverse transcription-polymerase chain reaction (RT-PCR) and western blot were carried out to detect ZSCAN31 expression in HCC cells. The results revealed that ZSCAN31 mRNA/protein expression was significantly decreased after TAZ knockdown (Figure 3A). Furthermore, ZSCAN31mRNA expression was higher in the HCC tissue samples than in the peritumoral tissues (Figure 3B). Figure 3C shows the results of immunostaining of HCC tissues. Compared with peritumoral tissue, ZSCAN31 was mainly expressed in the nucleus of HCC cells. This expression location was consistent with that of TAZ (9). Meanwhile, the up-regulation of ZSCAN31 was associated with poor OS (Figure 4A). As shown in Table 1, a significant difference was found in the 5-year OS rates of patients with low (48.8%) and high (32.0%) expression of ZSCAN31 (P=0.043).


Full table
Correlations between ZSCAN31 expression and the clinicopathological characteristics of HCC patients
Table 2 shows the clinical characteristics of the 83 HCC patients in our study. As with the expression of TAZ in our previous study, ZSCAN31 expression was significantly correlated with tumor volume (P<0.000), but not with portal vein emboli or metastasis (P=0.287). ZSCAN31 was highly expressed in the tissue samples of 50 patients (60.2%).

Full table
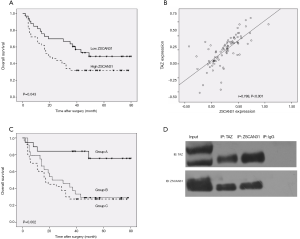
Coexpression of TAZ and ZSCAN31 has a better prognostic value for HCC after surgical resection
The expression levels of TAZ and ZSCAN31 in the 83 clinical HCC samples were further analyzed. A positive correlation was identified between TAZ and ZSCAN31 (r=0.788, P<0.01) (Figure 4B). Tumors tissue samples were divided into three groups based on the expression levels of TAZ and ZSCAN31 (Figure 4C). Group A (n=19) had low expression of TAZ and ZSCAN31; group B (n=24) had high expression TAZ or ZSCAN31; in group C (n=40), both TAZ and ZSCAN31 were highly expressed. Group A patients had a better OS rate than patients in group B and C (P=0.007) (Figure 4C, Table 1). Co-IP was used to detect the functional relevance of the interaction between TAZ and ZSCAN31. TAZ/ZSCAN31 was observed in complex with ZSCAN31/TAZ (Figure 4D). Taken together, our results show that the co-expression of TAZ and ZSCAN31 is a powerful predictor of poor prognosis after HCC surgical resection.
Discussion
As one of the nuclear effectors of the Hippo-related signaling pathways, the TAZ protein regulates cancer stem cell properties and the control of organ size (4,13-16). These biological processes have been observed in liver tumors, including HCC and ICC. In this study, ZSCAN31 was found to be down-regulated after TAZ knockdown, and high expression of ZSCAN31 was an indicator of poor OS in HCC patients. Additionally, a positive association was identified between the expressions of ZSCAN31 and TAZ in HCC, and Co-IP confirmed the interactions between the two genes. These results prove that ZSCAN31 expression can be analyzed to inform risk classification and treatment decision-making in HCC.
ZSCAN31 (also called ZNF323) plays an active role in human embryonic development (17). ZSCAN31 expression has been detected in many human organs, suggesting that it might be related to the development of multiple embryonic organs, including the liver. As a zinc-finger protein containing an N-terminal acidic domain and a C-terminal domain, ZSCAN31 may be involved in transcriptional regulation. Transcription factors (TFs) are the main regulators of gene expression in normal organ development, while changes in their expression pattern or function play a role in oncogenesis.
Previous studies have revealed that zinc-finger TFs have crucial involvement in tumorigenesis. For instance, the average ZNF165 mRNA expression in HCC is 88-fold higher than that in paired noncancerous liver tissues (18).
ZSCAN31 is situated on the major histocompatibility complex (MHC) region. This region on chromosome 6 is known to regulate multiple tumors, including Hodgkin lymphoma, esophageal squamous cell carcinoma, and HCC (19-21). The human leukocyte antigen (HLA)-G is a MHC class Ib gene, located on chromosome 6. Variant expression of HLA-G has been found in hepatitis B and C virus infections (22,23). Moreover, numerous studies have confirmed that HLA-G is expressed at high levels in HCC, finding HLA-G to be enriched in HCC tumor tissues and negatively associated with OS and recurrence in the HCC patients (24,25). This evidence suggests that ZSCAN31 is likely to be the susceptibility gene of HCC.
In summary, the clinical significance of the human SCAN/C2H2 zinc-finger transcription factor and ZSCAN31 we have analyzed. The biological function and the mechanism of the ZSCAN31 gene in HCC need further exploration.
Conclusions
In this study, ZSCAN31 was observed to be down-regulated after TAZ knockdown. A high expression level of ZSCAN31 was also found to be a predictor of poor OS in HCC patients. Additionally, ZSCAN31 expression was positively associated with TAZ expression in HCC, and the interactions between the two genes were confirmed with Co-IP. This study is the first to report that ZSCAN31 is an oncogene, particularly in HCC.
Acknowledgments
Funding: This study was funded by The National Natural Science Foundation of China (Grant No. 81702408), and the Science Foundation for Fostering Talents of The First Affiliated Hospital of Chongqing Medical University (Grant No. PYJJ2017-08).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-6373
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-6373
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-6373). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Clinical Specimens Ethics Committee of the First Affiliated Hospital of Chongqing Medical University. Signed informed consent forms were obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- He J, Gu D, Wu X, et al. Major Causes of Death among Men and Women in China. N Engl J Med 2005;353:1124-34. [Crossref] [PubMed]
- Cheung TT. Management of hepatocellular carcinoma: from bench to bedside and beyond. Transl Gastroenterol Hepatol 2019;4:54. [Crossref] [PubMed]
- Kanai F, Marignani PA, Sarbassova D, et al. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J 2000;19:6778-91. [Crossref] [PubMed]
- Hong JH, Hwang ES, McManus MT, et al. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science 2005;309:1074-8. [Crossref] [PubMed]
- Cordenonsi M, Zanconato F, Azzolin L, et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell 2011;147:759-72. [Crossref] [PubMed]
- Hong W, Guan KL. The YAP and TAZ transcription co-activators: key downstream effectors of the mammalian Hippo pathway. Semin Cell Dev Biol 2012;23:785-93. [Crossref] [PubMed]
- Matteucci E, Maroni P, Luzzati A, et al. Bone metastatic process of breast cancer involves methylation state affecting E-cadherin expression through TAZ and WWOX nuclear effectors. Eur J Cancer 2013;49:231-44. [Crossref] [PubMed]
- Yang N, Morrison CD, Liu P, et al. TAZ induces growth factor-independent proliferation through activation of EGFR ligand amphiregulin. Cell Cycle 2012;11:2922-30. [Crossref] [PubMed]
- Xiao H, Jiang N, Zhou B, et al. TAZ regulates cell proliferation and epithelial-mesenchymal transition of human hepatocellular carcinoma. Cancer Sci 2015;106:151-9. [Crossref] [PubMed]
- Xiao H, Tong R, Yang B, et al. TAZ regulates cell proliferation and sensitivity to vitamin D3 in intrahepatic cholangiocarcinoma. Cancer Lett 2016;381:370-9. [Crossref] [PubMed]
- Xiao H, Cheng S, Tong R, et al. BAG3 regulates epithelial-mesenchymal transition and angiogenesis in human hepatocellular carcinoma. Lab Invest 2014;94:252-61. [Crossref] [PubMed]
- Xiao H, Tong R, Ding C, et al. gamma-H2AX promotes hepatocellular carcinoma angiogenesis via EGFR/HIF-1alpha/VEGF pathways under hypoxic condition. Oncotarget 2015;6:2180-92. [Crossref] [PubMed]
- Lei QY, Zhang H, Zhao B, et al. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol Cell Biol 2008;28:2426-36. [Crossref] [PubMed]
- Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol 2011;13:877-83. [Crossref] [PubMed]
- Johnson R, Halder G. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat Rev Drug Discov 2014;13:63-79. [Crossref] [PubMed]
- Guo L, Teng L. YAP/TAZ for cancer therapy: opportunities and challenges Int J Oncol 2015;46:1444-52. (review). [Crossref] [PubMed]
- Pi H, Li Y, Zhu C, et al. A novel human SCAN/(Cys)2(His)2 zinc-finger transcription factor ZNF323 in early human embryonic development. Biochem Biophys Res Commun 2002;296:206-13. [Crossref] [PubMed]
- Dong XY, Yang XA, Wang YD, et al. Zinc-finger protein ZNF165 is a novel cancer-testis antigen capable of eliciting antibody response in hepatocellular carcinoma patients. Br J Cancer 2004;91:1566-70. [Crossref] [PubMed]
- Catamo E, Zupin L, Crovella S, et al. Non-classical MHC-I human leukocyte antigen (HLA-G) in hepatotropic viral infections and in hepatocellular carcinoma. Hum Immunol 2014;75:1225-31. [Crossref] [PubMed]
- Delahaye-Sourdeix M, Urayama KY, Gaborieau V, et al. A Novel Risk Locus at 6p21.3 for Epstein-Barr Virus-Positive Hodgkin Lymphoma. Cancer Epidemiol Biomarkers Prev 2015;24:1838-43. [Crossref] [PubMed]
- Zhao X, Sun Q, Tian H, et al. Loss of heterozygosity at 6p21 and HLA class I expression in esophageal squamous cell carcinomas in China. Asian Pac J Cancer Prev 2011;12:2741-5. [PubMed]
- Wang L, Zou ZQ, Wang K. Clinical Relevance of HLA Gene Variants in HBV Infection. J Immunol Res 2016;2016:9069375. [Crossref] [PubMed]
- Singh R, Kaul R, Kaul A, et al. A comparative review of HLA associations with hepatitis B and C viral infections across global populations. World J Gastroenterol 2007;13:1770-87. [Crossref] [PubMed]
- Cai MY, Xu YF, Qiu SJ, et al. Human leukocyte antigen-G protein expression is an unfavorable prognostic predictor of hepatocellular carcinoma following curative resection. Clin Cancer Res 2009;15:4686. [Crossref] [PubMed]
- Lin A, Chen HX, Zhu CC, et al. Aberrant human leucocyte antigen-G expression and its clinical relevance in hepatocellular carcinoma. J Cell Mol Med 2010;14:2162-71. [Crossref] [PubMed]
(English Language Editor: J. Reynolds)


