
Cimetidine promotes STUB1-mediated degradation of tumoral FOXP3 by activating PI3K-Akt pathway in gastric cancer
Introduction
Despite the comprehensive advances that have been achieved in the treatment of gastric cancer (GC) over the past few decades (1), the 5-year postoperative survival for patients with advanced GC remains low, with many patients still succumbing to GC metastasis and recurrence (2). Therefore, novel therapeutic options and strategies are required for GC therapy, especially for advanced or unresectable GC.
Histamine type-2 (H2) receptor is a promising and novel target for cancer therapy. Cimetidine, a type of H2 receptor antagonist, has been widely used in clinic to treat patients with peptic ulcers, acid reflux, and hypersecretory states (3). Cimetidine has also demonstrated antitumor effects on various types of malignancies, including colorectal cancer, renal cell carcinoma, malignant melanoma, glioblastoma, and GC (4-10). In one in vitro study, cimetidine inhibited the proliferation and induced apoptosis in colorectal cancer cells (11) and GC cells (12). Cimetidine has also been found to block the adhesion of colorectal tumor cells to endothelial cells via down-regulation of E-selectin on endothelial cells (13). Clinically, the postoperative administration of cimetidine at a normal therapeutic dosage was shown to significantly improve survival in GC patients of all stages (10). Similar beneficial effects have been reported in colorectal cancer (4-6). For instance, one study observed that cimetidine could increase the survival of colorectal cancer patients with high levels of sialyl Lewis antigens (ligand for E-selectin) (14), suggesting that E-selectin maybe a therapeutic target of cimetidine in colorectal cancer. Unfortunately, the therapeutic targets and the underlying mechanisms of cimetidine against GC are still unknown, and this limits the application of cimetidine in the clinical treatment of GC.Forkhead box protein 3 (FOXP3) is a member of the forkhead/winged helix family of transcription factors, which was previously believed to be restricted to regulatory T cells (Treg). It was reported that FOXP3 regulate diverse biological processes in Treg cells, including proliferation, differentiation, and functional regulation (15-18). Recent studies have identified the expression of endogenous FOXP3 in many solid tumors, including those of the breast (19), ovaries (20), lung (21), and prostate (22). We previously reported (23) that tumoral FOXP3 in GC cells could promote tumor proliferation, invasion, and migration, and that FOXP3 levels positively correlate with the malignancy of human GC tissues. Furthermore, prognostic analysis based on an online database (http://kmplot.com/analysis/) revealed that GC patients with a high level of FOXP3 expression showed a relatively shorter overall survival and progress-free survival than those with a low level of FOXP3 expression. These results indicate that tumoral FOXP3 may play a vital role in GC development. Therefore, further studies are required to understand how endogenous FOXP3 is regulated in GC development.
Recent studies have shown that stress, such as heat shock and lipopolysaccharide (LPS) stimulation promotes the downregulation of FOXP3 in Treg cells (24). Thus, cimetidine as a drug is also a kind of stress inducer, which may target endogenous FOXP3 in cancer, leading to tumor suppression. We speculated that cimetidine suppresses GC development by regulating tumoral FOXP3. This study is the first to report on the regulatory role and underlying mechanism of cimetidine on endogenous FOXP3 in GC cells. These findings may lead to the development of novel targeted therapeutic strategies against GC. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-6070).
Methods
Cell culture
Human GC cell strains (HGC27, SGC7901, MKN45, BGC823, and MGC803) and human normal gastric epithelial cell (GES-1) were maintained in RPMI-1640 medium (Gibco, USA), supplemented with 10% fetal bovine serum (FBS) (Hyclone, USA) and 1% penicillin/streptomycin (Gibco, USA) at 37°C and 5% CO2 in a humidified incubator. For the experiments, cells were treated with cimetidine (MedChem Express, cat. no. HY-14289) at a concentration of 4 mM, MG132 proteasome inhibitor (MedChem Express, cat. no. HY-13259) at a concentration of 10 µM, cycloheximide protein synthesis inhibitor (CHX) (MedChem Express, cat. no. HY-12320) at a concentration of 5 µg/mL, and LY294002 PI3K inhibitor (MedChem Express, cat. no. HY-10108) at a concentration of 5 and 10 µM.
Real-time polymerase chain reaction (RT-PCR)
Total RNA was extracted from cells using the TRIzol reagent (Applied Science, Germany) according to manufacturer’s instructions. The cDNA was then synthesized using the PrimeScript RT Reagent Kit (Takara, Japan) according to the manufacturer’s instructions. Real-time polymerase chain reaction (RT-PCR) was performed on the Applied Biosystems 7500 Real-Time PCR System (ABI, USA) with SYBR Premix Ex Taq (Takara, Japan) as previously reported. Relative expression level of target mRNAs was normalized to that of the endogenous control (β-actin), and was calculated based on the 2−∆∆CT comparative method according to the following formulae: ∆CT = (CTtarget − CTβCactin); and 2−∆∆CT = 2− (∆CTobservation group − ∆CTcontrol group). PCR for each sample was performed in triplicates. All primers used were as follows: FOXP3 forward: TACTTCAAGTTCCACAACATGCGACC; FOXP3 reverse: CGCACAAAGCACTTGTGCAGACTCAG; STUB1 forward: AGCAGGGCAATCGTCTGTTC; STUB1 reverse: CAAGGCCCGGTTGGTGTAATA; β-actin forward: CATGTACGTTGCTATCCAGGC; β-actin reverse: CTCCTTAATGTCACGCACGAT.
RNA interference
All small interfering RNAs (siRNAs) were purchased from GenePharma (Shanghai, China). Cells were transfected with 100 pmol of siRNA transiently at a cell confluency of 30–50% using siRNA-Mate (GenePharma, Shanghai, China) as previously reported (23). The sequences of siRNA duplexes are listed as follows: STUB1 forward: CCAGCGCUCUUCGAAUCGCGAAGAA; STUB1 reverse: UUCUUCGCGAUUCGAAGAGCGCUGG; NC forward: UUCUCCGAACGUGUCACGUTT; NC reverse: ACGUGACACGUUCGGAGAATT).
Western blotting
Total protein was extracted from cell homogenates and then immunoblotted with the following antibodies at 4 °C overnight: mouse mAb against human FOXP3 (236A/E7, Abcam, USA) at 1:200, rabbit pAb against human FOXP3 (GTX107737, GeneTex, USA) at 1:800, rabbit mAb against human STUB1 (EPR4447, Abcam, USA) at 1:500, rabbit mAb against ubiquitin (EPR8830, Abcam, USA)at 1:1,000, rabbit mAb against K48-linked ubiquitin (EP8589, Abcam, USA) at 1:1,000, rabbit mAb against Akt(pan) (#4685, CST, USA) at 1:1,000, rabbit mAb against pAkt(S473) (#4060, CST, USA) at 1:1,000, and rabbit mAb against pAkt(T308) (no.13038, CST, USA) at 1:1,000. After the final wash, membranes were incubated with horseradish peroxidase-conjugated secondary immunoglobin G (IgG) antibody (Jackson ImmunoResearch Laboratories, Inc., PA, USA) (1:10,000) at room temperature for 1 h. Membranes were developed using the Enhanced Chemiluminescence Detection Kit (Millipore, Germany).
CCK8 assay
Cells (5×103/well) were seeded into 96-well plates in triplicates and were cultured in 100 µL RPMI-1640 containing 10% FBS. The Cell Counting Kit-8 (CCK8) was used to evaluate cell proliferation. Briefly, 10 µL CCK8 solution was added to each plate, and cells were incubated for 2 h at 37 °C. OD450 was measured, andthe cell viability was calculated according to the formula: (experimental OD 450 value/control OD 450 value) ×100%.
Plate clone assay
Cells were seeded into 6-well plates with 100 or 500 cells in each well and were cultured in RPMI-1640 with 10% FBS. Upon visualizing the colonies after 10–14 days, the plates were washed with phosphate-buffered saline (PBS) and stained with crystal violet for 20 min. All experiments were performed in triplicate.
Matrigel invasion assay and cell migration assay
Transwell chambers with polyvinylidene fluoride (PVDF) filters (pore size 8.0 µm) were either uncoated (migration assay) or coated (invasion assay) with 50 µL diluted Matrigel. Cells (5×103–2×104/well) were suspended in 200 µL serum-free RPMI-1640, and seeded into the upper chambers in triplicates. RPMI-1640 containing 10% FBS (600 µL) was added to the lower chamber. Following a 24h incubation with or without cimetidine at 37 °C, the non-invading/migrating cells on the upper surface were removed. Cells that had migrated and adhered to the lower surface of the filter were fixed, stained with 0.1% crystal violet, and imaged. Cells in five non-overlapping fields were counted under a microscope (Olympus, Japan).
Wound healing assay
Cells (3×105/well) were seeded and grown in 6-well plates for 48 h until full confluence was reached. Wells were then washed with PBS, and a sterile 1,000 µL pipette tip was used to make a uniform scratch. After a 24-h incubation with or without cimetidine in serum-free RPMI-1640, the horizontal distance between the sides of the wound was measured. The experiments were performed in triplicate.
Immunoprecipitation (IP) and co-IP assay
Cells (5×106/well) were seeded and grown in 10-cm cell dishes for 24–48 h until 80% confluence was reached. Cells were then washed with PBS, and fresh medium with or without cimeditine was added for a12h incubation. Then, MG132 was added for another 6–8h incubation. Cells were harvested using IP lysis buffer, and centrifuged at >13,000 ×g for 30 min at 4°C, and the supernatants were collected for use. The IP and Co-IP were performed following the manufacturer’s instructions (PierceCrosslink Magnetic IP/Co-IP Kit, Thermo Scientific, cat. no. 88805). Briefly, the magnetic beads were washed with modified IP wash/lysis buffer, and FOXP3-specific antibody and IgG (negative control) were added to the magnetic beads for a ligation reaction (5 µg for each reaction). After the beads were washed with modified IP wash/lysis buffer, disuccinimidyl suberate (DSS) was added to strengthen the ligation between the beads and antibody to avoid the elution of antibody. Then, the cell lysates were added to the beads, and the mixture was incubated for 2 h at room temperature on a rotating platform. Finally, the beads were washed, and the FOXP3 complex was eluted using elution buffer.
Statistical analysis
Statistical analysis was performed using the SPSS20.0 statistical package. Quantitative data are presented as mean ± SD (standard deviation) and were analyzed by analysis of variance (ANOVA) or two-tailed Student’s t-tests. Categorical data were evaluated by the Chi-square test. A P value <0.05 was considered to be statistically significant.
Results
Cimetidine suppressed the proliferation, migration, and invasion of GC cells
In this study, we chose SGC7901 and HGC27 as the GC cell models, because the expressional level of FOXP3 was relatively lower in SGC7901 and higher in HGC27 in our previous study (23). GC cells and GES-1 were treated with cimetidine for 24 h at different concentrations (0, 0.25, 0.5, 1, 2, 4, and 8 mM) to screen the optimal dose. As shown in Figure 1A, the viability of GC cells decreased in a dose-dependent manner, while the viability of GES-1 began to decrease until 8 mM. Furthermore, the dose of 8mM began to cause a morphological change to GES-1 (data not shown). Hence, the dose of 4 mM was used as the experimental dose of cimetidine. Our study showed that cimetidine disrupted the proliferative capacity of GC cells by CCK8 (Figure 1B) and plate clone assay (Figure 1C). Both wound healing (Figure 1D) and transwell assay (Figure 1E) showed that cimetidine significantly reduced the number of migrated GC cells. In addition, Matrigel-based transwell assay (Figure 1F) demonstrated that cimetidine significantly reduced the number of invaded GC cells.
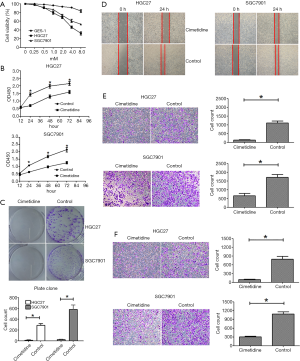
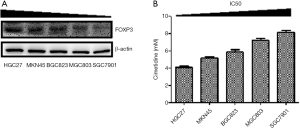
Interestingly, the antitumor effects of cimetidine on SGC7901 were less obvious than those on HGC27, especially in migratory and invasive capacities. We then analyzed the association between FOXP3 level and the sensitivity to cimetidine in a series of GC cell strains, including HGC27, MKN45, SGC7901, BGC823, and MGC803. As shown in Figure 2, CCK8 assay revealed that the IC50 of cimetidine inhibiting the viability of GC cells was increased with the decrease of FOXP3 level in these GC cells.
Cimetidine reduced the protein level of FOXP3 by proteasome-mediated ubiquitination/degradation
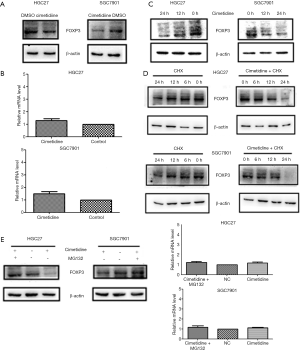
Our immunoblotting results showed that there was a decline in the protein level of FOXP3 in GC cells after a 24 h treatment of cimetidine (Figure 3A), but the mRNA level of FOXP3 increased slightly (Figure 3B). Further analysis showed the protein level of FOXP3 declined in a time-dependent manner (Figure 3C). Then, the half-life of FOXP3 protein was analyzed by adding protein synthesis inhibitor (CHX). It was observed that after a 12h-CHX treatment, the FOXP3 protein level was reduced more in cimetidine-treated GC cells than in untreated cells (Figure 3D). Then, proteasome inhibitor (MG132) was added in the cultured cells following 6–8 h of stimulation with cimetidine. We found that the addition of MG132 rescued the decline of the FOXP3 protein induced by cimetidine, while the mRNA level of FOXP3 was not significantly affected (Figure 3E).
STUB1 mediated the protein level of endogenous FOXP3
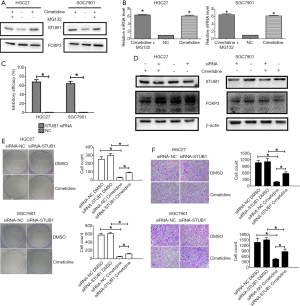
STUB1 is a kind of ubiquitin ligase E3, is widely expressed in tissues and cells, and participates in the ubiquitin modification of many protein substrates. We thus examined the effects of cimetidine on STUB1 in GC cells. Our results showed that cimetidine upregulated STUB1 at both the mRNA and protein level, while the addition of MG132 had no significant effects on it (Figure 4A,B). Then, we knocked down STUB1 in GC cells by using STUB1-specific siRNA (Figure 4C). It was noted that the knockdown of STUB1 rescued the decline of the FOXP3 protein in cimetidine-treated GC cells, while it had a minimal effect in untreated GC cells (Figure 4D). We then further examined the malignant behaviors of GC cells after the knockdown of STUB1 (Figure 4E,F); the knockdown of STUB1 reduced the turnover effect of cimetidine on proliferative and migratory capacities in GC cells, but had minimal effects on untreated GC cells.
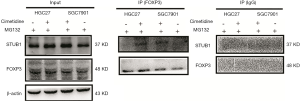
To explain this phenomenon, GC cells were treated with dimethyl sulfoxide (DMSO) or cimetidine, and MG132 was added after 6–8 h culture. FOXP3 immunoprecipitates in cells were extracted, and immunoblotted with anti-STUB1 antibody. As shown in Figure 5, STUB1 was only detected in FOXP3 immunoprecipitates in cimetidine-treated GC cells, but not in untreated GC cells.
Cimetidine upregulated STUB1 by activating PI3K/Akt pathway, leading to the degradation ofFOXP3 in GC cells
PI3K/Akt pathway is closely associated with tumor development. As shown in Figure 6A, cimetidine activated PI3K/Akt pathway in both GC cells. Furthermore, cimetidine mainly increased the phosphorylation of Akt (pAkt) level mainly at the T308 site in HGC27 cells and the S473 site in SGC7901 cells, while it had no effects on pan-Akt.
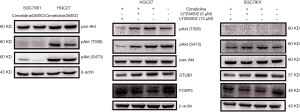
We then examined how PI3K/Akt pathway regulates the antitumor effects of cimetidine. The PI3K inhibitor (LY294002) was used to inhibit the phosphorylation of Akt, and we found that PI3K inhibitor can decrease the phosphorylation level of Akt at a concentration-dependent manner in both GC cells. Meanwhile, the inhibition of PI3K/Akt pathway partially rescued the increase of STUB1 induced by cimetidine. Furthermore, the inhibition of PI3K/Akt pathway also rescued the degradation of FOXP3 induced by cimetidine (Figure 6B).
Discussion
Cimetidine has been demonstrated to have the antitumor effects in GC patients. Tonnesen et al. were the first to report that cimetidine improved survival in GC patients (10). Some studies have shown that cimetidine modulates the apoptotic pathway in GC cells. Furthermore, other H2 receptor antagonists, such as ranitidine and famotidine, do not demonstrate such effects (25,26), implying that the antitumor effects of cimetidine are unique. It is widely believed that cimetidine can enhance immune surveillance against tumor cells by suppressing immune suppressor cells, including Treg cells by targeting FOXP3 (4,27-29), negate the effect of histamine on tumor growth and tumor angiogenesis (7,25,30,31), and suppress cell adhesion (13). However, the therapeutic targets and regulatory mechanisms related to these processes remain elusive.
In this study, we found that cimetidine inhibited the proliferative, migratory, and invasive capacities of GC cells. These results were consistent with the previous findings (10), which found that cimetidine could suppress GC cell growth and induce GC cell apoptosis; our study further confirmed and detailed the beneficial effects of cimetidine on GC cells. Furthermore, we also found that GC cells with higher FOXP3 were more susceptible to cimetidine treatment. This indicates that FOXP3 might be a therapeutic target of cimetidine in GC, and that the endogenous FOXP3 level may affect the sensitivity to cimetidine treatment in GC cells.
We then focused on determining whether and how cimetidine regulates the endogenous FOXP3 in GC cells. We found that cimetidine downregulated the protein level of FOXP3, but had no effect on mRNA level. Further study indicated that cimetidine substantially reduced the half-life of FOXP3 protein, indicating cimetidine may downregulate FOXP3 protein at a post-translational level. The proteasome system regulates the ubiquitination/degradation of target proteins, which is an important way to control protein quantity and quality at a post-translational level. Thus, MG132 was used to inhibit the role of proteasome, and we found that MG132 rescued the decline of FOXP3 protein induced by cimetidine, indicating cimetidine might downregulate FOXP3 protein by promoting the ubiquitination/degradation of FOXP3.
It has been reported that environmental stress can induce protein damage and destabilization, and these damaged or unstable proteins are targeted for ubiquitination/degradation to prevent accumulation in cells (32,33). Cimetidine stimulation can be also considered as a kind of environmental stress and may destabilize or damage the endogenous FOXP3 protein in tumor cells, initiating the process of ubiquitination/degradation. Three kinds of enzymes, ubiquitin-activating enzyme E1, ubiquitin-transferase E2, and ubiquitin ligase E3, participate in the process of ubiquitination. Ubiquitin ligase E3 is substrate-specific and considered as the most important enzyme. STUB1, also named CHIP (the C-terminus of Hsp70-interacting protein), is a U-box-type ubiquitin ligase E3, which is an important member of ubiquitin-proteasome systems, and mediates the degradation of substrates, including many oncogenic proteins. STUB1 has been shown to produce all known types of ubiquitination modifications of the substrates (34), which then can be degraded by the 26S proteasome (35,36). A previous study (24) showed that STUB1 is the ubiquitin ligase E3 that regulates the ubiquitination/degradation of FOXP3 in Treg cells. We consequently examined whether STUB1 participates in cimetidine-induced degradation of FOXP3 in GC cells. Our study showed that cimetidine promoted the transcription and translation of STUB1 and that the knockdown of STUB1 rescued the decline of FOXP3 protein in cimetidine-treated GC cells, but had no significant effect in untreated cells. In addition, the knockdown of STUB1 had a minimal effect on the malignant behaviors of GC cells. Furthermore, when the GC cells were exposed to cimetidine, the knockdown of STUB1 partially rescued the turnover effect of cimetidine on GC cells, including proliferation and migration. These results suggested that STUB1-mediated degradation of endogenous FOXP3 in GC cells was cimetidine stimulus-dependent. We further examined the STUB1 level in FOXP3 precipitates extracted from cimetidine-treated and untreated cells. It was found that STUB1 was only detected in FOXP3 immunoprecipitates from cimetidine-treated cells, indicating that cimetidine promoted the formation of FOXP3-STUB1 complex, thereby leading to the ubiquitination/degradation of FOXP3.
In our study, we also found that PI3K/Akt pathway participated in the process of cimetidine against GC. Cimetidine promoted the transcription and translation of STUB1 by activating PI3K/Akt pathway, and then STUB1 further promoted the degradation of FOXP3. The findings of this study reinforced our understanding of PI3K/Akt pathway in tumor. This pathway may play a dual (pro- or anti-) role in tumor development, and is affected by multiple factors, such as the environment, trigger factors, downstream targets, etc. Here, we concluded that cimetidine promotes STUB1-mediated ubiquitination/degradation of tumoral FOXP3 by activating PI3K/Akt pathway, leading to GC suppression (Figure 7).
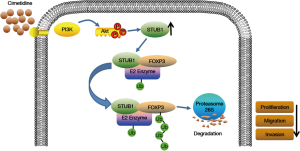
To date, few data on the regulatory mechanism of FOXP3 in tumor have been published. To our knowledge, this is the first study to report that cimetidine promotes STUB1-mediated ubiquitination/degradation of FOXP3 by activating PI3K/Akt pathway, thereby suppressing tumor development in GC. Identifying the therapeutic target of cimetidine against cancer is significant, as it may facilitate the screening of patients sensitive to cimetidine treatment and improve the therapeutic efficacy. The role of FOXP3 in Treg cells has been clarified. Cimetidine may regulate cancer itself and the tumor microenvironment if it targets FOXP3 in both Treg cells and cancer cells. However, further studies are required to confirm this hypothesis.
The emergence of targeted therapy drugs has greatly promoted the progress of gastric cancer treatment, but tumor cells inevitably develop drug resistance, which limits the application in clinical practice. We think more molecular targets need to be found to solve this problem, which can form a target panel. And a combined medicine targeting a panel of molecular targets may reduce the occurrence of drug resistance.
Some limitations in this study should be addressed. First, our study could not exclude other E3 ubiquitin ligases that may mediate the ubiquitination of FOXP3 protein in GC cells, because one protein can be regulated by several E3 ubiquitin ligases. Second, the types of ubiquitination modifications of FOXP3 were not examined, but our study indirectly indicated a K48-ubiquitination modification of FOXP3, which will need to been confirmed in the next study. Third, no animal study was performed to examine the role of cimetidine on GC in vivo, due to the establishment of spontaneous gastric cancer mouse is as long as 12 months, which will be our next research plan. Finally, FOXP3 maybe a sensitive therapeutic target of cimetidine in the treatment of GC, but this conclusion cannot be verified in GC cell lines, and further, more in-depth study is needed. For example, by using ideal cell models such as organoids, the sensitive target or the genetic features associated with the sensitivity to cimetidine treatment can be identified.
To conclude, our study re-emphasized the antitumor effect of cimetidine, and indicated that tumoral FOXP3 may be a sensitive therapeutic target candidate of cimetidine in GC. This finding expands our understanding of the therapeutic mechanism of cimetidine. Cimetidine has been used clinically for many years, and it is generally safe and well tolerated, making cimetidine a valuable additional treatment for many types of cancer. With the identification of therapeutic and sensitive targets, we can screen sensitive cancer patients and selectively administer cimetidine treatment concomitantly with antitumor drugs or other therapeutic strategies. This is a promising path in optimizing the current therapeutic strategies and for improving the clinical efficacy in the treatment of advanced GC.
Acknowledgments
Funding: This work was partially supported by the National Natural Science Foundation of China (No. 81802996, No. 81871946), the Primary Research & Development Plan of Jiangsu Province (No. BE2016786), the Jiangsu Key Medical Discipline (General Surgery) (No. ZDXKA2016005), the youth foundation of the Jiangsu Province Health Bureau (No. BK20161062), the Program for Development of Innovative Research Team in the First Affiliated Hospital of NJMU, the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD, JX10231801), and the Jiangsu Key Lab of Cancer Biomarkers, Prevention and Treatment, Jiangsu Collaborative Innovation Center for Cancer Personalized Medicine.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-6070
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-6070
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-6070). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Padilla-Leal KE, Medina-Franco H. Eighth edition of the American Joint Committee on Cancer staging system: are we getting closer to the ideal classification for gastric cancer? Ann Transl Med 2019;7:S52.
- Shinde A, Novak J, Amini A, et al. The evolving role of radiation therapy for resectable and unresectable gastric cancer. Transl Gastroenterol Hepatol 2019;4:64. [Crossref] [PubMed]
- Welage LS. Overview of pharmacologic agents for acid suppression in critically ill patients. Am J Health Syst Pharm 2005;62:S4-10. [Crossref] [PubMed]
- Adams WJ, Morris DL. Short-course cimetidine and survival with colorectal cancer. Lancet 1994;344:1768-9. [Crossref] [PubMed]
- Morris DL, Adams WJ. Cimetidine and colorectal cancer--old drug, new use? Nat Med 1995;1:1243-4. [Crossref] [PubMed]
- Kelly MD, King J, Cherian M, et al. Randomized trial of preoperative cimetidine in patients with colorectal carcinoma with quantitative assessment of tumor-associated lymphocytes. Cancer 1999;85:1658-63. [Crossref] [PubMed]
- Dexeus FH, Logothetis CJ, Sella A, et al. Phase II study of coumarin and cimetidine in patients with metastatic renal cell carcinoma. J Clin Oncol 1990;8:325-9. [Crossref] [PubMed]
- Morton RF, Creagan ET, Cullinan SA, et al. Phase II studies of single-agent cimetidine and the combination N-phosphonacetyl-L-aspartate (NSC-224131) plus L-alanosine (NSC-153353) in advanced malignant melanoma. J Clin Oncol 1987;5:1078-82. [Crossref] [PubMed]
- Lefranc F, Yeaton P, Brotchi J, et al. Cimetidine, an unexpected anti-tumor agent, and its potential for the treatment of glioblastoma Int J Oncol 2006;28:1021-30. (review). [Crossref] [PubMed]
- Tonnesen H, Knigge U, Bulow S, et al. Effect of cimetidine on survival after gastric cancer. Lancet 1988;2:990-2. [Crossref] [PubMed]
- Rajendra S, Mulcahy H, Patchett S, et al. The effect of H2 antagonists on proliferation and apoptosis in human colorectal cancer cell lines. Dig Dis Sci 2004;49:1634-40. [Crossref] [PubMed]
- Jiang CG, Liu FR, Yu M, et al. Cimetidine induces apoptosis in gastric cancer cells in vitro and inhibits tumor growth in vivo. Oncol Rep 2010;23:693-700. [PubMed]
- Kobayashi K, Matsumoto S, Morishima T, et al. Cimetidine inhibits cancer cell adhesion to endothelial cells and prevents metastasis by blocking E-selectin expression. Cancer Res 2000;60:3978-84. [PubMed]
- Matsumoto S, Imaeda Y, Umemoto S, et al. Cimetidine increases survival of colorectal cancer patients with high levels of sialyl Lewis-X and sialyl Lewis-A epitope expression on tumour cells. Br J Cancer 2002;86:161-7. [Crossref] [PubMed]
- Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science 2003;299:1057-61. [Crossref] [PubMed]
- Hori S, Sakaguchi S. Foxp3: a critical regulator of the development and function of regulatory T cells. Microbes Infect 2004;6:745-51. [Crossref] [PubMed]
- Sakaguchi S, Ono M, Setoguchi R, et al. Foxp3+ CD25+ CD4+ natural regulatory T cells in dominant self-tolerance and autoimmune disease. Immunol Rev 2006;212:8-27. [Crossref] [PubMed]
- Lin Z, Liu X, Liu X, et al. DNA hypermethylation of the promoter attenuates forkhead box protein 3 (FOXP3) expression in hepatocellular carcinoma cells. Transl Cancer Res 2019;8:2024-31. [Crossref]
- Zuo T, Wang L, Morrison C, et al. FOXP3 is an X-linked breast cancer suppressor gene and an important repressor of the HER-2/ErbB2 oncogene. Cell 2007;129:1275-86. [Crossref] [PubMed]
- Zhang HY, Sun H. Up-regulation of Foxp3 inhibits cell proliferation, migration and invasion in epithelial ovarian cancer. Cancer Lett 2010;287:91-7. [Crossref] [PubMed]
- Chen GY, Chen C, Wang L, et al. Cutting edge: Broad expression of the FoxP3 locus in epithelial cells: a caution against early interpretation of fatal inflammatory diseases following in vivo depletion of FoxP3-expressing cells. J Immunol 2008;180:5163-6. [Crossref] [PubMed]
- Wang L, Liu R, Li W, et al. Somatic single hits inactivate the X-linked tumor suppressor FOXP3 in the prostate. Cancer Cell 2009;16:336-46. [Crossref] [PubMed]
- Zhang L, Xu J, Zhang X, et al. The Role of Tumoral FOXP3 on Cell Proliferation, Migration, and Invasion in Gastric Cancer. Cell Physiol Biochem 2017;42:1739-54. [Crossref] [PubMed]
- Chen Z, Barbi J, Bu S, et al. The ubiquitin ligase Stub1 negatively modulates regulatory T cell suppressive activity by promoting degradation of the transcription factor Foxp3. Immunity 2013;39:272-85. [Crossref] [PubMed]
- Lawson JA, Adams WJ, Morris DL. Ranitidine and cimetidine differ in their in vitro and in vivo effects on human colonic cancer growth. Br J Cancer 1996;73:872-6. [Crossref] [PubMed]
- Hahm KB, Kim WH, Lee SI, et al. Comparison of immunomodulative effects of the histamine-2 receptor antagonists cimetidine, ranitidine, and famotidine on peripheral blood mononuclear cells in gastric cancer patients. Scand J Gastroenterol 1995;30:265-71. [Crossref] [PubMed]
- Kumar A, Cleveland RP. Immunoregulatory effects of cimetidine: inhibition of suppressor cell effector function in vivo. Immunopharmacol Immunotoxicol 1988;10:327-32. [Crossref] [PubMed]
- Adams WJ, Morris DL, Ross WB, et al. Cimetidine preserves non-specific immune function after colonic resection for cancer. Aust N Z J Surg 1994;64:847-52. [Crossref] [PubMed]
- Sahasrabudhe DM, McCune CS, O'Donnell RW, et al. Inhibition of suppressor T lymphocytes (Ts) by cimetidine. J Immunol 1987;138:2760-3. [PubMed]
- Reynolds JL, Akhter J, Morris DL. In vitro effect of histamine and histamine H1 and H2 receptor antagonists on cellular proliferation of human malignant melanoma cell lines. Melanoma Res 1996;6:95-9. [Crossref] [PubMed]
- Natori T, Sata M, Nagai R, et al. Cimetidine inhibits angiogenesis and suppresses tumor growth. Biomed Pharmacother 2005;59:56-60. [Crossref] [PubMed]
- Burkewitz K, Choe K, Strange K. Hypertonic stress induces rapid and widespread protein damage in C. elegans. Am J Physiol Cell Physiol 2011;301:C566-76. [Crossref] [PubMed]
- Pratt WB, Morishima Y, Peng HM, et al. Proposal for a role of the Hsp90/Hsp70-based chaperone machinery in making triage decisions when proteins undergo oxidative and toxic damage. Exp Biol Med (Maywood) 2010;235:278-89. [Crossref] [PubMed]
- Kim HT, Kim KP, Lledias F, et al. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem 2007;282:17375-86. [Crossref] [PubMed]
- McDonough H, Patterson C. CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones 2003;8:303-8. [Crossref] [PubMed]
- Hatakeyama S, Yada M, Matsumoto M, et al. U box proteins as a new family of ubiquitin-protein ligases. J Biol Chem 2001;276:33111-20. [Crossref] [PubMed]
(English Language Editor: J. Gray)


