
Stromal cells promote chemoresistance of acute myeloid leukemia cells via activation of the IL-6/STAT3/OXPHOS axis
Introduction
Acute myeloid leukemia (AML) is a heterogeneous group of hematologic malignancies that differ in biological characteristics, clinical course, and prognosis. In recent years, considerable progress has been made in the diagnosis and treatment of leukemia. However, although the majority of AML patients initially respond to chemotherapy, over 50% of the cases relapsed presenting with drug resistance and poor prognosis (1). Increasing evidence suggests that interactions between leukemia cells and surrounding bone marrow stromal cells (BMSCs) provide favorable conditions for AML cells to evade chemotherapy (2). Our previous study also demonstrated that co-culturing with BMSCs protected AML cells from daunorubicin (DNR), homoharringtonine, and cytosine arabinoside (Ara-C), by upregulating the PI3K/Akt signaling pathway (3).
BMSCs secrete a variety of cytokines and growth factors into microenvironment. Interleukin (IL)-6, IL-8, IFN-γ, and TNF-α, are well known to be involved in regulation of tumorigenesis and cancer progression by affecting invasion, proliferation, migration, and apoptosis of tumor cells (2,4-6). The IL-6 family of cytokines activate the signal transducer and activator of transcription 3 (STAT3), a transcription factor known to promote tumorigenesis (7-9). Increased levels of IL-6 in plasma have been shown to inversely correlate with disease-free survival rates in patients with AML (10). Twenty-five percent of the patients with AML were reported to constitutively express phosphorylated STAT3 due to high IL-6 secretion levels (11). Both autocrine and paracrine IL-6 can lead to constitutive phosphorylation of STAT3 at Tyr705 and Ser727 residues; IL-6 from paracrine sources (such as cancer-associated myeloid cells or fibroblasts) could induce autocrine IL-6 production and STAT3 phosphorylation in tumor cells (9). Owing to the association between increased STAT3 activity and chemotherapeutic resistance and poor prognosis in AML (11,12), a variety of therapeutic strategies have been developed to target STAT3. For instance, C188-9, a high-affinity STAT3 inhibitor, was shown to prevent granulocyte colony-stimulating factor-induced STAT3 phosphorylation and induce apoptosis in AML cells (13). Besides functioning as a transcription factor in cancer, serine phosphorylated STAT3 (pS-STAT3) can also localize in the mitochondria, where it modulates mitochondrial oxidative phosphorylation (OXPHOS) via interaction with components of the mitochondrial electron transport chain (9,14-17). Upregulation of mitochondrial STAT3 and OXPHOS are the assumed mechanisms of resistance to tyrosine kinase inhibitors in cancer stem cells undergoing metabolic reprogramming (17).
Mitochondria are metabolic hubs that integrate diverse mechanisms to provide energy for the cell, and aberrant function of these cellular organelles is implicated in the etiology and pathological consequences of several diseases (18). Cancer cells reprogram their metabolism to satisfy the needs of enhanced growth, proliferation and long-term persistence in the organism (19,20). Contrary to the Warburg theory, which argues that tumorigenesis is promoted by insufficient respiration due to mitochondrial dysfunction, recent data suggest that in various tumor types including breast, cervical, pancreatic and liver cancers, energy is provided by switching the metabolic activity to mitochondrial OXPHOS (21,22). Furthermore, upregulation of OXPHOS may contribute to chemoresistance of cancer cells (23,24). AML cells have been shown to heavily depend on OXPHOS (25,26), and thus, a small-molecule inhibitor of OXPHOS, IACS-010759, could efficiently reduce AML cell proliferation and induce apoptosis through energy depletion and decreased production of aspartate (27). Similarly, the STAT3 inhibitor, OPB-111077, has anticancer effects in patients with diffuse large B-cell lymphoma through OXPHOS blockage (28). These observations suggest a relationship among STAT3 signaling, OXPHOS, and tumorigenesis in AML model. However, little is known about the role of OXPHOS in BMSC-mediated AML chemoresistance.
In the present study, we report that BMSCs alter mitochondrial OXPHOS in AML cells, which contributes to chemoresistance in AML cells likely through the activation of the IL-6/STAT3/OXPHOS axis. The induction of OXPHOS in AML cells caused by BMSCs may be a potential therapeutic target to enhance the response of patients with AML to chemotherapy and prevent disease recurrence.
We present the following article in accordance with the Materials Design Analysis Reporting (MDAR) checklist (available at http://dx.doi.org/10.21037/atm-20-3191).
Methods
Cell lines and primary AML cells
Human AML cell lines HL-60, U-937 and THP-1, were purchased from Cell Bank of Type Culture Collection Chinese Academy of Sciences (Shanghai, China) and cultured in RPMI 1640 medium (Hyclone, Logan, UT, USA) containing 10% fetal bovine serum (FBS, Gibco, Life Technologies, Grand Island, NY, USA), 2 mM L-glutamine (Gibco), 100 units/mL of penicillin, and 100 µg/mL of streptomycin (Gibco). Human BMSCs cell lines HS-5 were purchased from American Type Culture Collection (Manassas, VA, USA) and cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Hyclone) supplemented with 10% FBS, 2 mM L-glutamine, 100 units/mL of penicillin, and 100 µg/mL of streptomycin (Gibco). Cells were maintained in a humidified 5% CO2 incubator at 37 °C.
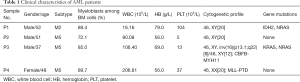
Primary AML cells were isolated from newly diagnosed AML patients before any therapeutic intervention at the Fujian Medical University Union Hospital; patients’ clinical information is presented in Table 1. BM aspirates of AML patients were processed by Ficoll density gradient centrifugation to isolate mononuclear cells followed by CD34+ selection according to the manufacturer’s instructions (Miltenyi Biotec, Shanghai, China). Primary AML cells were cultured in the same conditions as AML cell lines. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of the Fujian Medical University Union Hospital (No. 2019KJCX029) and informed consent was taken from all the patients.
Full table
Transwell co-culture system
To avoid a direct cell-cell contact, we adopted the transwell co-culture system. HS-5 cells were plated at a density of 1×105 cells/well in the lower compartment of a 6-well transwell plate (CoStar, Cambridge, MA, USA) and cultured until 70–80% confluence; then, 4×105 HL-60, U-937, THP-1 cells, or primary AML cells were seeded onto the permeable polycarbonate membrane insert of 0.4-µm pore size (CoStar) while sharing the culture medium. Monocultures of HS-5 or AML cells were used as controls. Cells were harvested after 24 h.
Mitochondrial function and glycolytic rate assays
To analyze the mitochondrial function and glycolytic rate, the mitochondrial oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were determined using the Seahorse XF Cell Mito Stress Test Kit and the Seahorse XF Glycolytic Rate Assay Kit (Agilent Technologies), respectively, and analyzed in a Seahorse XFe24 cell metabolic analyzer (Agilent Technologies). Briefly, AML cells (2×105/well) were allowed to adhere to XFe24 cell culture microplates (Agilent Technologies) using Cell-Tak (BD Biosciences) and incubated at 37 °C for 1 h before analysis. The sensor cartridge in Seahorse XF Calibrant was hydrated at 37 °C in a non-CO2 incubator overnight. In the cell mitochondrial stress test, oligomycin (1 µM), carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone (FCCP, 2 µM), and rotenone and antimycin A (Rot/AA, 0.5 µM) were injected sequentially at specific time points. Oligomycin is an inhibitor of ATP synthase (complex V) and decreases the OCR through affecting ATP production. FCCP is a mitochondrial OXPHOS inhibitor used to examine spare respiratory capacity indicative of the cellular potential to respond to increased energy demands. Rot and AA are complex I and III inhibitors, respectively, which in combination block mitochondrial respiration, thereby enabling the assessment of non-mitochondrial respiration. In the glycolytic rate assay, Rot/AA (0.5 µM) and an inhibitor of glycolysis 2-deoxy-D-glucose (2-DG, 50 mM) were added sequentially at the indicated time points.
Cell viability assay
The sensitivity of AML cells to chemotherapeutic agents was determined as previously described (29). Briefly, AML cells were plated in 96-well plates in growth medium and treated with 200 ng/mL DNR (Xinshidai, Shandong, China), 10 µM Ara-C (Cytosar, Foshan, China), or PBS (vehicle control), and cytotoxicity was assessed after 24 h by a cell viability assay (Cell Counting Kit-8, CCK-8, Dojindo Laboratories, Kumamoto, Japan).
RNA-seq and pathway enrichment analysis
Total RNA was extracted from 1×106 HL-60 or U-937 cells cultured with or without HS-5 cells, and its concentration and integrity were measured using the Nanodrop One (Thermo Fisher Scientific Inc., Carlsbad, CA, USA) and the Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). Preparation of the cDNA library from 1 µg RNA consisted of the following steps: mRNA enrichment with oligo (dT)-containing magnetic beads, cDNA synthesis and purification, repair of DNA ends, addition of a poly-A tail and adaptor, selection of the appropriate fragment size, and PCR enrichment. The obtained library was checked for quality using the Agilent Bioanalyzer 2100 system and sequenced on an Illumina HiSeq2500 platform. After sequencing, the raw data were processed to obtain clean data by removing reads containing adapter or poly-N sequences and those of low quality. Differences in gene expression between sample groups were analyzed by the DESeq software package and the gene list was then created according to −log10 (Q-value) ranking. Pathway enrichment analysis of the ranked gene list was conducted using the gene enrichment analysis (GSEA) (http://software.broadinstitute.org/gsea/) (30). Calculations were performed based on three independent biological replicates.
Real-time fluorescence quantitative PCR (RT-qPCR)
Total RNA was extracted with the TRIzol reagent (Invitrogen; Thermo Fisher Scientific) and cDNA was synthesized using the Revert Aid First Strand cDNA Synthesis Kit (Invitrogen) according to the manufacturer’s instructions. RT-qPCR was carried out in an ABI 7500 instrument (Thermo Fisher Scientific, Singapore, Singapore) using FastStart Universal SYBR Green Master (Rox) (Roche Applied Science, Basel, Switzerland) with primers specific for IL-6 (forward 5'-CCCCTCAGCAATGTTGTTTGT-3' and reverse 5'-CTCCGGGACTGCTAACTGG-3'), IL-6R (forward 5'-TTCTACAGACTACGGTTTGAG-3' and reverse 5'-GGATGACACAGTGATGCT-3'), IL-6ST (forward 5'-ACTGTTGATTATTCTACTGTGTAT-3' and reverse 5'-AATTATGTGGCGGATTGG-3'), β-actin (forward 5'-CTGGGACGACATGGAGAAAA-3' and reverse 5'-AAGGAAGGCTGGAAGAGTGC-3') and GAPDH (forward 5'-AGAAGGCTGGGGCTCATTTG-3' and reverse 5'-AGGGGCCATCCACAGTCTTC-3'). The reaction was performed at the following cycling conditions: denaturation at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The relative mRNA expression was calculated after normalization with β-actin or GAPDH levels using the 2−ΔΔCt method (31).
Detection of STAT3 activity
The STAT3 activator, colivelin (CLN), and the STAT3 inhibitor, C188-9, were purchased from Med Chem Express (Monmouth Junction, NJ, USA). AML cells were treated individually with 50 ng/mL IL-6, 100 nM CLN, or 10 µM C188-9 for 24 h. Next, AML cells were co-cultured with HS-5 cells for 24 h and then treated with 10 µM C188-9 for 24 h. To assess total STAT3 activity, AML cells were collected, fixed in 4% formaldehyde in PBS for 15 min at 25 °C, washed with PBS, and permeabilized in chilled 90% methanol for 30 min on ice. Cells were then incubated with Alexa Fluor 488 Mouse Anti-STAT3 (pY705 or pS727) antibody (BD Biosciences) for 30 min at room temperature and analyzed by flow cytometry (Accuri C6 Plus, BD Biosciences, Franklin Lakes, NJ, USA). The data were processed using the FlowJo software Version 10.6.0. To assess mitochondrial STAT3 activity, the mitochondria of AML cells were extracted using a cell mitochondria isolation kit (Beyotime Institute of Biotechnology, Nantong, China) according to the manufacturer’s instructions. Mitochondrial extracts were evaluated by Western blot analysis.
Western blot analysis
Cellular and mitochondrial extracts were used to perform Western blot analysis as previously described (32) using the following antibodies: human IL-6 rabbit monoclonal antibody, STAT3 rabbit monoclonal antibody, phospho-STAT3 (pS727) rabbit monoclonal antibody, COX IV rabbit monoclonal antibody (1:1,000; Cell Signaling Technology, Danvers, MA, USA), β-actin or GAPDH mouse monoclonal antibody (1:1,000; Beyotime Institute of Biotechnology), and horseradish peroxidase (HRP)-conjugated goat anti-mouse or rabbit IgG (H+L) (1:5,000; Beyotime Institute of Biotechnology). Chemiluminescence signals were visualized using the BeyoECL Star kit (Beyotime Institute of Biotechnology) and detected with the ChemiDoc Touch system (Bio-Rad, Hercules, CA, USA). Quantification of band intensities was carried out using the ImageJ software.
Production of lentivirus particles and infection
Single guide (sg) RNAs were designed using the MIT CRISPR design software (http://crispr.mit.edu) and oligonucleotides including sgRNA sequences (IL-6 exon 1, 5'-ACCCCCAATAAATATAGGAC-3') were synthesized by Sangon Biotech (Shanghai, China). LentiCRISPRv2 was a gift from Brett Stringer (RRID: Addgene_98290; http://n2t.net/addgene:98290). Lentiviral vectors containing IL-6-targeting sgRNAs were generated as described previously (33,34), verified by DNA sequencing, and used to transfect 293T cells. Non-targeting empty vector LentiCRISPRv2 was used as a negative control. HS-5 cells were infected with the produced lentiviral particles. Stable IL-6 knockout (IL-6KO) or control (IL-6KO-Con) cells were selected in the presence of puromycin (0.25 µg/mL; d) for 12 days, and the efficiency of IL-6 knockout was validated by immunoblotting and RT-qPCR.
Enzyme linked immunosorbent assay (ELISA)
Quantitation of IL-6 secretion by HS-5 cells or AML cells was determined using the human IL-6 ELISA Kit (CUSABIO, Wuhan, China) according to the manufacturer’s instructions.
Measurement of intracellular reactive oxygen species (ROS) level
Total intracellular ROS was determined by staining cells with the cell-permeable fluorogenic probe dichlorofluorescein diacetate (DCFH-DA, Beyotime) that would become fluorescent when DCFH was oxidized to fluorescein by intracellular ROS. Briefly, cells were washed with PBS, incubated with 10 µM DCFH-DA at 37 °C for 30 min, washed three times with PBS, and assessed by flow cytometry (C6 plus, BD Biosciences).
Statistical analysis
The data are expressed as the mean ± standard deviation of three independent experiments. All analyses were performed using GraphPad Prism version 6.05 (GraphPad Software Inc., La Jolla, CA, USA). The significance of differences between groups was determined using the Student’s t-test; P<0.05 was considered statistically significant.
Results
BMSCs promoted chemoresistance in AML cells by upregulating OXPHOS
Aerobic glycolysis is generally associated with the proliferation of cancer cells (20), and OXPHOS has recently been shown to be upregulated in certain cancers including leukemia (21,22). To address the role of these metabolic processes in AML chemoresistance induced by HS-5 cells, we measured aerobic glycolysis and OXPHOS using the XFe24 cell metabolic analyzer. Co-culture with HS-5 cells significantly stimulated mitochondrial respiration in AML cells, as evidenced by an increase in the levels of basal respiration and ATP production (Figure 1A,B), but did not affect aerobic glycolysis (Figure 1C). We also measured maximal respiration and spare respiratory capacity, two main indicators of the cell ability to respond to energy demands. The results showed that AML cells co-cultured with HS-5 cells also increased maximal and spare respiration to meet metabolic demands (Figure 1A,B). Additionally, the co-cultured AML cells that had exhibited higher OXPHOS also produced more ROS than the mono-cultured AML cells (Figure 1D). Furthermore, HL-60, U-937, and THP-1 cells co-cultured with HS-5 cells increased resistance to chemotherapeutic drugs DNR and Ara-C, compared with monocultures (Figure 1E), suggesting that HS-5 cells protected AML cells from drug-induced toxic effects. To determine whether the increased chemoresistance in co-cultured AML cells was related to the induction of OXPHOS, we used a low concentration of oligomycin A, which inhibited mitochondrial respiration but did not affect the survival of AML cells. The results showed that treatment with oligomycin A significantly increased chemosensitivity in co-cultured AML cells (Figure 1E), suggesting that the chemoresistance in AML cells induced by HS-5 cells was mediated partially by upregulating OXPHOS in AML cells.

BMSCs activated IL-6/STAT3 signaling and mitochondrial STAT3 in AML cells
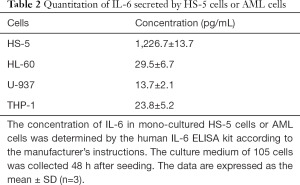
RNA-seq and a pathway enrichment analysis were used to explore the molecular mechanism underlying upregulation of OXPHOS in AML cells induced by BMSCs. Results from the GSEA indicated that the IL-6/STAT3 signaling cascade was significantly enriched in HL-60 and U-937 cells after co-cultured with HS-5 cells (Figure 2A). We detected the concentration of IL-6 in the culture medium of monocultured HS-5 or AML cells using ELISA. A substantial amount of IL-6 (approximately 1,200 pg/mL) was secreted into the culture medium by HS-5 cells within 48 h (Table 2) whereas AML cells secreted less than 30 pg/mL within 48 h, accounting for about a 40-fold difference. Additionally, we used RT-qPCR to analyze the expression of IL-6 receptor IL-6R and its binding partner gp130 in HL-60 and U-937 cells after co-culture with HS-5 cells. mRNA expression levels of the IL-6R gene and the gp130-encoding IL-6ST gene were significantly upregulated in co-cultured AML cells when compared with those in monocultures (Figure 2B). Furthermore, the phosphorylation levels of STAT3 (both pY705 and pS727) was increased in AML cells after being co-cultured with HS-5 cells or treated with IL-6 (Figure 2C), suggesting that BMSCs activated the IL-6/STAT3 pathway in AML cells. These results reflected the transcriptional function of STAT3 in AML cells located in the marrow microenvironment.

Full table
According to previous reports that implicate the activation of mitochondrial STAT3 (pS727) as a key link between cellular signaling pathways and cancer cell metabolic activity [14-17], we proposed that HS-5 induced upregulation of mitochondrial OXPHOS might be interrelated with the activity of STAT3 in mitochondria in AML cells. Therefore, we extracted mitochondria from co-cultured AML cells and performed Western blot analysis to assess pS-STAT3 level in mitochondria, with β-actin and histone H3 employed as indicators of cytoplasmic and nuclear protein contamination, respectively. Co-cultured AML cells showed significant increases of pS-STAT3 level in mitochondria (Figure 2D), thereby indicating a role for BMSCs in activating mitochondrial STAT3 in AML cells.
BMSCs upregulated the level of OXPHOS in AML cells by activating the mitochondrial STAT3
To test whether phosphorylation of the mitochondrial STAT3 was required for BMSCs to upregulate the OXPHOS in AML cells, AML cells were treated with STAT3 phosphorylation activator (CLN) or inhibitor (C188-9). Treatment with CLN was effective to activate the total and the mitochondrial STAT3 phosphorylation in monocultured AML cells (Figure 3A,B,C,D). C188-9 was bound to the SH2 domain of STAT3, and decreased STAT3 tyrosine phosphorylation (13). Our results showed that C188-9 decreased total and mitochondrial pS-STAT3 levels in both monocultured and co-cultured AML cells as well (Figure 3A,B,C,D). Mitochondrial function analysis showed that the basal or maximal respiration, spare respiratory capacity, and ATP production were increased in CLN-treated AML cells, suggesting that CLN increased OXPHOS levels of AML cells (Figure 4A). C188-9 inhibited OXPHOS and decreased the cellular ability to respond to metabolic demands in both mono- and co-cultured AML cells, as evidenced by decreasing levels of basal and maximal respiration, spare respiratory capacity, and ATP production (Figure 4A,B,C). Notably, C188-9 enhanced chemosensitivity of AML cells after co-culturing, but CLN increased chemoresistance in monocultured AML cells (Figure 4D). These results indicated that BMSCs upregulated OXPHOS levels in AML cells by activating the mitochondrial STAT3, thereby promoting chemoresistance in AML cells.


BMSCs activated mitochondrial STAT3 in AML cells by secreting more IL-6
As intercellular effects were reciprocal, it is likely that AML cells in turn could affect the biological activity of BMSCs. Indeed, our data demonstrated that HS-5 cells co-cultured with AML cells had significantly higher mRNA and protein expression of IL-6 than HS-5 monocultures (Figure 5A). Given this observation, we thus speculated that IL-6 released from HS-5 cells activated mitochondrial STAT3 in AML cells. We used the CRISPR/Cas9 system to knock out IL-6 from HS-5 cells, and examined the phosphorylation status of STAT3 in AML cells co-cultured with the IL-6 knockout HS-5 (HS-5/IL-6KO) cells. As shown in Figure 5B, the sgRNA was effective in reducing IL-6 expression at the protein level. AML cells co-cultured with HS-5/IL-6KO cells not only had lower phosphorylation levels of total STAT3 (Figure 5C), but also showed a decrease in mitochondrial pS-STAT3 levels (Figure 5D). In contrast, treatment with extraneous IL-6 increased both total and mitochondrial STAT3 activity in AML cells (Figure 5C,D). Thus, BMSCs activated mitochondrial STAT3 in AML cells by inducing IL-6 secretion.

IL-6 secreted by BMSCs enhanced mitochondrial OXPHOS in AML cells to confer chemoresistance
Our results showed that OXPHOS upregulated in AML cells by BMSCs was linked to mitochondrial STAT3 activation, which led us to assess the effect of IL-6 secreted by BMSCs on OXPHOS in AML cells. Consistently, the levels of OXPHOS, basal and maximal respiration, spare respiratory capacity as well as ATP production were significantly elevated in HS-5 co-cultured AML cells, whereas knockout of IL-60 in HS-5 cells abolished these effects (Figure 6A,B). Extraneous IL-6 alone could enhance OXPHOS and metabolic fitness in monocultures of AML cells, accompanied by increased chemoresistance (Figure 6A,B). Co-culture with HS-5/IL-6KO cells rendered AML cells hypersensitive to the chemotherapeutic drugs as compared to the AML cells co-cultured with HS-5/IL-6 KO-Con cells (Figure 6C). Similar to the results obtained with AML cell lines, all patient-derived AML cells became resistant when co-cultured with HS-5/IL-6KO-Con cells, but gained sensitivity when co-cultured with HS-5/IL-6KO cells (Figure 6D). Thus, the data is most consistent with the concept, as pictorially modelled in Figure 7, that AML cells induced BMSCs to release more IL-6, which in turn upregulated OXPHOS in co-cultured AML cells, thus enhancing chemoresistance in AML cells, including primary AML cells.


Discussion
Residence of AML cells within the BM niche is considered an important factor of enhancing their chemoresistance because leukemia cells may remodel the microenvironment to support their survival and division (2,35,36). AML cells are exposed to a variety of paracrine signals from BMSCs that alter the behavior of AML cells, ultimately promoting disease progression (2,36). IL-6 secreted by BMSCs has been shown to contribute to the development of several cancers such as multiple myeloma (37), lung cancer (38), hepatocellular carcinoma (39), and breast cancer (40). High levels of IL-6 in serum of patients with AML and chronic lymphocytic leukemia were associated with more aggressive disease, although serum IL-6 could originate from other sources apart from stromal cells (41,42). In this study, we revealed that IL-6 secreted by BMSCs altered mitochondria OXPHOS in AML cells through the activation of the STAT3 signaling, which enhanced chemoresistance of AML cells. Additionally, our results confirmed that AML cells stimulated BMSCs to secrete more IL-6, which in turn activated the total and mitochondrial STAT3 in AML cells, thereby promoting proliferation. The IL-6/STAT3 pathway has been shown to contribute to the development of several cancers, including AML (9,42) and abnormal activation of STAT3 is known to be associated with poor prognosis. Thus, activation of the IL-6/STAT3 pathway was more frequently detected in high-risk cancers (43) and was shown to increase chemoresistance in AML (12). Therefore, blocking IL-6 with specific antibodies (e.g., siltuximab) or targeting STAT3 by small-molecule inhibitors (e.g., C188-9) have been tested as new therapeutic approaches for AML (10,13,44). However, these studies were mostly focused on the role of IL-6/STAT3 signaling in inducing proliferation and inhibiting apoptosis of AML cells, and the mechanism of IL-6/STAT3 signaling in the chemoresistance of AML cells induced by BMSCs in the marrow microenvironment is far from clear.
Previous reports have shown that mitochondrial STAT3 interacts with GRIM-19 (component of complexes I of the mitochondrial electron transport chain) to modulate OXPHOS (9,14-17). Our study demonstrated that IL-6/STAT3 signaling played an important role in the BMSC-induced enhancement of the mitochondrial OXPHOS in AML cells, thereby leading to chemoresistance of AML cells. Accumulating evidence suggests that tumor cells develop resistance to chemotherapy by shifting their energy metabolism towards OXPHOS (21,22,24). A recent study showed that human AML cells with high rather than low levels of OXPHOS were chemoresistant in vivo and that a metabolic shift toward lower OXPHOS significantly enhanced antileukemic effects of Ara-C (45), which was consistent with our findings. Our study is the first to demonstrate that OXPHOS in AML cells was closely associated with the bone marrow (BM) microenvironment; BMSCs enhanced mitochondrial OXPHOS and increased chemoresistance in AML cells via IL-6/STAT3 signaling. Additionally, our results further support the notion that activation of mitochondrial STAT3 is a key link between cellular signaling pathways and cancer cell metabolic activity (14-17,46). Inhibition of OXPHOS in tumor cells through downregulation of STAT3 has proved to be effective in preclinical studies (17,28,47). Further identification of biomarkers associated with downregulation of OXPHOS and discovery of inhibitors with acceptable toxicity profiles holds promise for overcoming chemoresistance in AML and other cancers.
It is noteworthy that this study only probes the IL-6 secreted by BMSCs. First, AML cells are exposed to a variety of paracrine signals from BMSCs that alter the behavior of AML cells, ultimately promoting the progression of cancer (2,36). Second, endogenous cytokines significantly affect the response of cells to tumor microenvironmental stimuli. For instance, IL-6 from paracrine sources may induce autocrine production of IL-6 and STAT3 phosphorylation in tumor cells (9). Lastly, there are significant individual differences in cytokine profiles observed in the BM microenvironment. Thus, the association between cytokine levels and clinical outcomes should be further evaluated (10,42). Furthermore, the transcriptional and mitochondrial effects of p-STAT3 occur almost simultaneously. The role of STAT3 as a nuclear transcription factor is considerable, as evidenced by the RNA-seq and flow cytometry analysis results. However, our research focused on the role of mitochondrial function in microenvironment-induced AML chemoresistance, and our results suggested that the mitochondria play an important role. In addition, it should also be recognized that only primary bone marrow mesenchymal stem cell (BMMSC) can fulfill stringent in vivo stem cell criteria. Thus, further studies to determine the magnitude and quality of primary BMMSC on upregulation of OXPHOS in AML cells to facilitate chemoresistance are clearly warranted.
Tumor cell metabolism has been considered as a hallmark of cancer, and tumor cells adapt cellular metabolism to cope with their high energy requirement by switching between glycolysis and OXPHOS. We found that aerobic glycolysis was not affected in the co-cultured AML cells (Figure 1C), indicating that IL-6 from BMMSC did not affect AML cells through the “Warburg effect”. Ždralević et al. (48) reported and discussed the potential therapeutic benefit of disrupting the Warburg effect, which could re-route cancer cells to OXPHOS thus offering a vulnerability point via ferroptosis-induced cell death. Ferroptosis, a rather novel cell death mechanism, is driven by the failure of glutathione-dependent antioxidant defenses and subsequent iron-dependent accumulation of lipid ROS (49). ROS are produced as a byproduct intracellularly by mitochondrial OXPHOS. Both this study and the one by Farge et al. (45) showed that Ara-C-resistant AML cells exhibited high-OXPHOS status and produced high levels of ROS. A recent study has also indicated that high-OXPHOS metabolism could enhance chemo-sensitivity by coordinating ROS levels in human ovarian cancers, which is mediated by ferroptosis (50). Therefore, modulation of ferroptosis may be another potential anticancer strategy for AML.
Conclusions
In summary, our data suggest that OXPHOS upregulation in AML cells by BMSCs may be a potential therapeutic target to overcome chemoresistance and prevent recurrence of AML. Although our results were obtained in vitro and need to be validated in animal models and clinical specimens, they add to the growing body of evidence that the control of cytokine production in BM and downregulation of OXPHOS in AML cells may be a viable approach for developing novel therapeutic strategies to enhance drug sensitivity in AML and improving patients’ outcome.
Acknowledgments
Funding: This work was supported by grants from the Startup Fund for Scientific Research at Fujian Medical University (2016QH034), the National Natural Science Foundation of China (81870108 and 81800136), the Fujian Medicine Innovation Program (2018-CX-18), Joint Funds for the Innovation of Science and Technology, Fujian Province (2017Y9053 and 2019Y9073), the Government-funded Project of the Construction of High-level Laboratory (201704), National Undergraduate Training Program for Innovation and Entrepreneurship (201910392011) and National and Fujian Provincial Key Clinical Specialty Discipline Construction Program, China.
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/atm-20-3191
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-3191
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-3191). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of the Fujian Medical University Union Hospital (No. 2019KJCX029) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Burnett A, Wetzler M, Lowenberg B. Therapeutic advances in acute myeloid leukemia. J Clin Oncol 2011;29:487-94. [Crossref] [PubMed]
- Duarte D, Hawkins ED, Lo Celso C. The interplay of leukemia cells and the bone marrow microenvironment. Blood 2018;131:1507-11. [Crossref] [PubMed]
- Chen P, Jin Q, Fu Q, et al. Induction of Multidrug Resistance of Acute Myeloid Leukemia Cells by Cocultured Stromal Cells via Upregulation of the PI3K/Akt Signaling Pathway. Oncol Res 2016;24:215-23. [Crossref] [PubMed]
- Kyurkchiev D, Bochev I, Ivanova-Todorova E, et al. Secretion of immunoregulatory cytokines by mesenchymal stem cells. World J Stem Cells 2014;6:552-70. [Crossref] [PubMed]
- Kumar R, Godavarthy PS, Krause DS. The bone marrow microenvironment in health and disease at a glance. J Cell Sci 2018;131:jcs201707. [Crossref] [PubMed]
- Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med 2013;19:1423-37. [Crossref] [PubMed]
- Heinrich PC, Behrmann I, Müller-Newen G, et al. Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem J 1998;334:297-314. [Crossref] [PubMed]
- Bollrath J, Phesse TJ, von Burstin VA, et al. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell 2009;15:91-102. [Crossref] [PubMed]
- Sansone P, Bromberg J. Targeting the interleukin-6/Jak/stat pathway in human malignancies. J Clin Oncol 2012;30:1005-14. [Crossref] [PubMed]
- Sanchez-Correa B, Bergua JM, Campos C, et al. Cytokine profiles in acute myeloid leukemia patients at diagnosis: survival is inversely correlated with IL-6 and directly correlated with IL-10 levels. Cytokine 2013;61:885-91. [Crossref] [PubMed]
- Zhou J, Bi C, Janakakumara JV, et al. Enhanced activation of STAT pathways and overexpression of survivin confer resistance to FLT3 inhibitors and could be therapeutic targets in AML. Blood 2009;113:4052-62. [Crossref] [PubMed]
- Benekli M, Xia Z, Donohue KA, et al. Constitutive activity of signal transducer and activator of transcription 3 protein in acute myeloid leukemia blasts is associated with short disease-free survival. Blood 2002;99:252-7. [Crossref] [PubMed]
- Redell MS, Ruiz MJ, Alonzo TA, et al. Stat3 signaling in acute myeloid leukemia: ligand-dependent and -independent activation and induction of apoptosis by a novel small-molecule Stat3 inhibitor. Blood 2011;117:5701-9. [Crossref] [PubMed]
- Wegrzyn J, Potla R, Chwae YJ, et al. Function of mitochondrial Stat3 in cellular respiration. Science 2009;323:793-7. [Crossref] [PubMed]
- Meier JA, Larner AC. Toward a new STATe: the role of STATs in mitochondrial function. Semin Immunol 2014;26:20-8. [Crossref] [PubMed]
- Yu H, Lee H, Herrmann A, et al. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer 2014;14:736-46. [Crossref] [PubMed]
- Lee M, Hirpara JL, Eu JQ, et al. Targeting STAT3 and oxidative phosphorylation in oncogene-addicted tumors. Redox Biol 2019;25:101073. [Crossref] [PubMed]
- Spinelli JB, Haigis MC. The multifaceted contributions of mitochondria to cellular metabolism. Nat Cell Biol 2018;20:745-54. [Crossref] [PubMed]
- Cantor JR, Sabatini DM. Cancer cell metabolism: one hallmark, many faces. Cancer Discov 2012;2:881-98. [Crossref] [PubMed]
- Liberti MV, Locasale JW. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem Sci 2016;41:211-8. [Crossref] [PubMed]
- Weinberg SE, Chandel NS. Targeting mitochondria metabolism for cancer therapy. Nat Chem Biol 2015;11:9-15. [Crossref] [PubMed]
- Pollak M. Targeting oxidative phosphorylation: why, when, and how. Cancer Cell 2013;23:263-4. [Crossref] [PubMed]
- Haq R, Shoag J, Andreu-Perez P, et al. Oncogenic BRAF regulates oxidative metabolism via PGC1alpha and MITF. Cancer Cell 2013;23:302-15. [Crossref] [PubMed]
- Hirpara J, Eu JQ, Tan JKM, et al. Metabolic reprogramming of oncogene-addicted cancer cells to OXPHOS as a mechanism of drug resistance. Redox Biol 2019;25:101076. [Crossref] [PubMed]
- Sriskanthadevan S, Jeyaraju DV, Chung TE, et al. AML cells have low spare reserve capacity in their respiratory chain that renders them susceptible to oxidative metabolic stress. Blood 2015;125:2120-30. [Crossref] [PubMed]
- Skrtic M, Sriskanthadevan S, Jhas B, et al. Inhibition of mitochondrial translation as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell 2011;20:674-88. [Crossref] [PubMed]
- Molina JR, Sun Y, Protopopova M, et al. An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nat Med 2018;24:1036-46. [Crossref] [PubMed]
- Tolcher A, Flaherty K, Shapiro GI, et al. A First-in-Human Phase I Study of OPB-111077, a Small-Molecule STAT3 and Oxidative Phosphorylation Inhibitor, in Patients with Advanced Cancers. Oncologist 2018;23:658-e72. [Crossref] [PubMed]
- Chen P, Wen X, Wang B, et al. PI3K/Akt inhibitor LY294002 potentiates homoharringtonine antimyeloma activity in myeloma cells adhered to stromal cells and in SCID mouse xenograft. Ann Hematol 2018;97:865-75. [Crossref] [PubMed]
- Reimand J, Isserlin R, Voisin V, et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat Protoc 2019;14:482-517. [Crossref] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Chen P, Zhan W, Wang B, et al. Homoharringtonine potentiates the antileukemic activity of arsenic trioxide against acute myeloid leukemia cells. Exp Cell Res 2019;376:114-23. [Crossref] [PubMed]
- Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 2014;11:783-4. [Crossref] [PubMed]
- Shalem O, Sanjana NE, Hartenian E, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014;343:84-7. [Crossref] [PubMed]
- Duan CW, Shi J, Chen J, et al. Leukemia propagating cells rebuild an evolving niche in response to therapy. Cancer Cell 2014;25:778-93. [Crossref] [PubMed]
- Mohammadi S, Nikbakht M, Sajjadi SM, et al. Reciprocal Interactions of Leukemic Cells with Bone Marrow Stromal Cells Promote Enrichment of Leukemic Stem Cell Compartments in Response to Curcumin and Daunorubicin. Asian Pac J Cancer Prev 2017;18:831-40. [PubMed]
- Wang J, De Veirman K, De Beule N, et al. The bone marrow microenvironment enhances multiple myeloma progression by exosome-mediated activation of myeloid-derived suppressor cells. Oncotarget 2015;6:43992-4004. [Crossref] [PubMed]
- Li L, Han R, Xiao H, et al. Metformin sensitizes EGFR-TKI-resistant human lung cancer cells in vitro and in vivo through inhibition of IL-6 signaling and EMT reversal. Clin Cancer Res 2014;20:2714-26. [Crossref] [PubMed]
- Mi F, Gong L. Secretion of interleukin-6 by bone marrow mesenchymal stem cells promotes metastasis in hepatocellular carcinoma. Biosci Rep 2017;37:BSR20170181. [Crossref] [PubMed]
- Hartman ZC, Poage GM, den Hollander P, et al. Growth of triple-negative breast cancer cells relies upon coordinate autocrine expression of the proinflammatory cytokines IL-6 and IL-8. Cancer Res 2013;73:3470-80. [Crossref] [PubMed]
- Yoon JY, Lafarge S, Dawe D, et al. Association of interleukin-6 and interleukin-8 with poor prognosis in elderly patients with chronic lymphocytic leukemia. Leuk Lymphoma 2012;53:1735-42. [Crossref] [PubMed]
- Stevens AM, Miller JM, Munoz JO, et al. Interleukin-6 levels predict event-free survival in pediatric AML and suggest a mechanism of chemotherapy resistance. Blood Adv 2017;1:1387-97. [Crossref] [PubMed]
- Marotta LL, Almendro V, Marusyk A, et al. The JAK2/STAT3 signaling pathway is required for growth of CD44(+)CD24(-) stem cell-like breast cancer cells in human tumors. J Clin Invest 2011;121:2723-35. [Crossref] [PubMed]
- Yao X, Huang J, Zhong H, et al. Targeting interleukin-6 in inflammatory autoimmune diseases and cancers. Pharmacol Ther 2014;141:125-39. [Crossref] [PubMed]
- Farge T, Saland E, de Toni F, et al. Chemotherapy-Resistant Human Acute Myeloid Leukemia Cells Are Not Enriched for Leukemic Stem Cells but Require Oxidative Metabolism. Cancer Discov 2017;7:716-35. [Crossref] [PubMed]
- Szczepanek K, Lesnefsky EJ, Larner AC. Multi-tasking: nuclear transcription factors with novel roles in the mitochondria. Trends Cell Biol 2012;22:429-37. [Crossref] [PubMed]
- Mackenzie GG, Huang L, Alston N, et al. Targeting mitochondrial STAT3 with the novel phospho-valproic acid (MDC-1112) inhibits pancreatic cancer growth in mice. PLoS One 2013;8:e61532. [Crossref] [PubMed]
- Ždralević M, Vučetić M, Daher B, et al. Disrupting the 'Warburg effect' re-routes cancer cells to OXPHOS offering a vulnerability point via 'ferroptosis'-induced cell death. Adv Biol Regul 2018;68:55-63. [Crossref] [PubMed]
- Yang WS, Stockwell BR. Ferroptosis: Death by Lipid Peroxidation. Trends Cell Biol 2016;26:165-76. [Crossref] [PubMed]
- Gentric G, Kieffer Y, Mieulet V, et al. PML-Regulated Mitochondrial Metabolism Enhances Chemosensitivity in Human Ovarian Cancers. Cell Metab 2019;29:156-173.e10. [Crossref] [PubMed]


