
A HPV16-related prognostic indicator for head and neck squamous cell carcinoma
Introduction
The human papillomavirus (HPV) is a major cause for infectious agent-related cancers, including cervical cancer and head and head and neck squamous cell carcinoma (HNSCC) (1). It contributes to an estimated 600,000 or more incident cancer cases and 250,000 deaths each year (2,3). In addition, an increasing number of studies have demonstrated superior prognosis and different genomic and phenotypic characteristics of patients with HPV-associated HNSCC (4-7).
MicroRNAs (miRNAs) play pivotal roles in post-transcriptional regulation of gene expression in HPV-related cancers (8,9). High-risk HPV16 type genes (E5, E6, E7) have been reported to interact with miRNAs in tumor progression (10-13). HPV+ HNSCC showed distinct miRNA profile. These miRNAs may potentially target common HPV-mediated pathways such as E6/p53 and E7/pRb (14,15). However, miRNA function on the prognosis of HPV16+ HNSCC was unclear.
This study aimed to identify potential miRNAs and their roles in HPV16+ HNSCC progression, based on the The Cancer Genome Atlas (TCGA) database. Potential target genes were predicted, and possible functions were also analyzed. We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/atm-20-6338).
Methods
Data processing
All clinical and sequencing data was obtained from The TCGA database (https://cancergenome.nih.gov/). Based on the inclusion criteria of cases with complete miRNA expression, mRNA expression and corresponding clinical data, 519 HNSCC patients and 44 corresponding HNSCC-negative control samples were included in this study. The raw data was converted into a data matrix in R, and the identification of differentially expressed miRNAs (DEMs) and mRNAs (DEGs) was performed in HNSCC and normal samples through the R package, Limma. In addition, DEMs between HNSCC patients with different HPV 16 statuses were also selected. All DEMs or DEGs meeting the cutoff criteria |log2(Fold Change)|>1 and P<0.05 were considered significant. Univariate Cox regression analysis was performed to select significant prognostic miRNAs. Common DEMs were identified using the plug-in jvenn (http://jvenn.toulouse.inra.fr/app/index.html) (16), and the final prognostic miRNAs were determined by multivariate Cox regression analysis. In addition, a risk model based on 3 miRNAs was built with a risk
Correlation of DEMs with clinical and pathological characteristics of HNSCC patients
The expression levels of selected DEMs with clinical and pathological parameters (gender, age, alcohol and smoking history, cancer grade, cancer stage, T-stage, N stage, HPV16 status, perineurial invasion (PNI), angiolymphatic invasion, disease surgical margin status) were compared using the unpaired t test or one-way ANOVA.
Association of DEMs and patient prognosis
The expression levels of selected DEMs underwent log2 transformation before analysis. Each patient was classified as having either positive or negative expression according to the corresponding median miRNA expression. The Kaplan-Meier approach with log-rank analysis using the R package (survival) was adopted to determine the role of candidate prognostic miRNAs in HNSCC. In addition, overall survival (OS) analysis for HNSCC patients with high or low risk, based on the calculated risk score was also performed to estimate the prognostic value of all selected DEMs.
Target gene prediction
Predicted target genes of the selected DEMs were verified using the miRDB database (17).
Gene Ontology (GO) functional and pathway enrichment analyses and Gene Set Enrichment Analysis (GSEA)
The GO functions with respect to the biological process (BP), molecular function (MF), and cellular component (CC) categories and KEGG pathway enrichment for the predicted target genes were identified through the Metascape database with a P<0.05 cutoff.
GSEA for DEGs in HPV16+ HNSCC patients [|log2(Fold Change)|>1 and P<0.05] was performed using R package clusterProfiler. Hallmark gene sets were selected for functional analysis.
Statistics analysis
Statistical analyses were performed using the R program. Analytical data is expressed as mean ± standard deviation (SD). Unpaired t-tests or one-way ANOVA were performed to assess the relationships between clinical and pathological characteristics, along with miRNA expression levels. Analysis of parameters with 2 groups was performed using an unpaired t-test, while analysis of parameters with 3 or more groups was performed using one-way ANOVA. Pearson correlation analysis was performed for each pair of miRNAs expressed in HNSCC samples. Univariate and multivariate Cox proportional hazard regression analyses and Kaplan-Meier survival analysis were conducted to compare the prognostic values of high- and low-level miRNAs. Differences with a P value <0.05 were considered significant.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Results
Determination of prognostic DEMs in HPV+ HNSCC
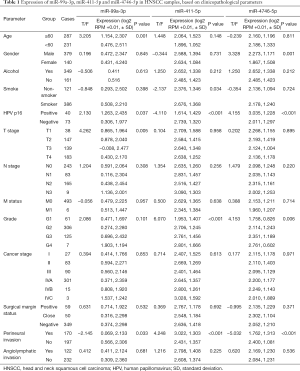
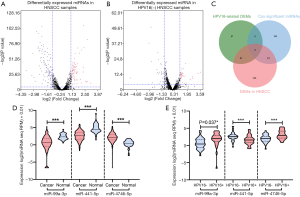
Table 1 lists the expression of miR-99a-3p, miR-411-5p and miR-4746-5p in HNSCC samples based on clinicopathological parameters. Based on the cutoff criteria (P<0.05 and |log2FC|>1.0), 183 DEMs were identified between cancer and normal samples. An additional 59 DEMs were found between HNSCC patients with varied HPV16 statuses (Figure 1). Furthermore, 195 potential prognostic miRNAs were selected by univariate analysis with the cutoff criterion of P<0.05. As shown in Figure 1C, 6 common miRNAs were selected (miR-99a-3p, miR-411-5p, miR-432-5p, miR-495-3p, miR-758-5p, miR-4746-5p). Subsequently, final prognostic miRNAs (miR-99a-3p, miR-411-5p, miR-4746-5p) were confirmed by multivariate Cox regression analysis with the cutoff P<0.05 (Table 2).
Full table

Full table
miR-99a-3p, miR-411-5p, and miR-4746-5p showed significant differential expression in HPV16+ HNSCC tumors
As shown in Figure 1D, both miR-99a-3p and miR-411-5p were frequently downregulated in HNSCC compared to normal samples (miR-99a-3p: 0.469±2.254 vs. 2.339±0.823, P<0.0001; miR-411-5p: 2.604±1.366 vs. 4.747±1.329, P<0.0001). The expression level of miR-4746-5p was significantly upregulated in HNSCC samples (2.159±1.204 vs. 0.368±1.019, P<0.0001). In HPV16+ HNSCC samples, miR-99a-3p and miR-4746-5p were significantly upregulated and miR-411-5p was downregulated [miR-99a-3p: HPV16(+) 1.263±2.435, HPV16(−) 0.306±1.977, P=0.037; miR-411-5p: HPV16(+) 1.614±1.429, HPV16(−) 2.739±1.320, P<0.001; miR-4746-5p: HPV16(+) 3.035±1.228, HPV16(−) 2.011±1.297, P<0.001, Figure 1E].
Expression levels of miR-99a-3p, miR-411-5p, and miR-4746-5p reflecting PNI status of HNSCC patients
As shown in Table 1, the expression levels of miR-99a-3p, miR-411-5p, and miR-4746-5p in HNSCC patients with different clinical and pathological characteristics were also analyzed. The expression of all 3 miRNAs was significantly different with regard to PNI status. In HNSCC patients with positive PNI, miR-99a-3p and miR-4746-5p were significantly downregulated and miR-411-5p was upregulated (miR-99a-3p, P=0.033; miR-411-5p, P<0.001; miR-4746-5p, P<0.001, Figure 1F).
Besides, miR-411-5p and miR-4746-5p showed significantly different expression levels in HNSCC patients with regard to cancer grade (miR-411-5p, P<0.001; miR-4746-5p, P=0.006), while miR-99a-3p showed significantly different expression in HNSCC patients with regard to cancer T stage (P=0.005). In addition, miR-99a-3p expression was upregulated in older HNSCC patients (≥60: 1.154±2.307, <60: 0.476±2.511, P=0.001). Furthermore, miR-411-5p expression was downregulated in HNSCC patients with a confirmed smoking history (smoker: 2.376±1.346, non-smoker: 2.676±1.368, P=0.034) and miR-4746-5p expression was upregulated in male HNSCC patients (male: 2.273±1.171, female: 1.867±1.508, P=0.001). All data are summarized in Table 1.
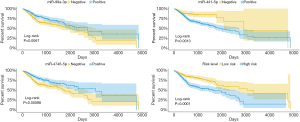
HNSCC patients with high miR-99a-3p and miR-4746-5p expression, along with low miR-411-5p expression displayed better OS
The OS of 518 HNSCC patients was analyzed in groups with different expression levels of miR-99a-3p, miR-411-5p and miR-4746-5p (Figure 2). Cox regression analysis (Table 2) revealed miR-411-5p as a risk factor (HR =1.756, P=0.043), and miR-99a-3p (HR=0.658, P=0.008) in addition to miR-4746-5p (HR =0.650, P=0.008) are protective factors in HNSCC. As shown in Figure 3, HNSCC patients with low miR-411-5p (P=0.0013) and high miR-99a-3p expression (P=0.0068), along with miR-4746 (P<0.001) exhibited significantly improved OS. Furthermore, a risk model was constructed, based on the risk score of miR-99a-3p, miR-411-5p and miR-4746-5p, determined by multivariate Cox regression analysis. All HNSCC samples were categorized into a high-risk or low-risk group, based in the median risk score. HNSCC patients in the high-risk group showed significantly poorer OS than patients in the low-risk group (P<0.0001).

Target gene prediction and functional analysis of miR-99a-3p, miR-411-5p, and miR-4746-5p
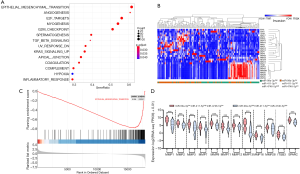
Three hundred and eighty-two predicted target genes of miR-99a-3p, miR-411-5p and miR-4746-5p were identified from the miRDB database. GO and KEGG pathway analysis were performed on Metascape with the cutoff P<0.05.
As shown in Figure 3, significant BP terms included regulation of cellular catabolic process, gamete generation, regulation of protein catabolic process, developmental cell growth, neuroepithelial cell differentiation and neuron projection extension. The MF and CC terms were mainly enriched in signal sequence binding and the axon, respectively. Moreover, KEGG pathways were enriched in the MAPK and FoxO signaling pathways, which suggested that target genes may be associated with cancer and neural growth.
Correlation between the downregulation of epithelial mesenchymal transition (EMT) genes in HNSCC patients with miR-99a-3phigh miR-411-5plow miR-4746-5phigh expression
To determine the possible effects of miR-99a-3p, miR411-5p and miR-4746-5p in HPV16+ HNSCC patients, we screened the DEGs in HNSCC samples with miR-99a-3phigh miR-411-5plow miR-4746-5phigh expression prior to undertaking GSEA analysis. Analysis of hallmark gene sets displayed significant changes of gene expression related to EMT, angiogenesis, E2F-targets and myogenesis in HNSCC patients with miR-99a-3phigh miR-411-5plow miR-4746-5phigh expression (Figure 4A). EMT gene sets were significantly downregulated in HNSCC patients with miR-99a-3phigh miR-411-5plow miR-4746-5phigh expression (Figure 4B).
Genes related to invasion were downregulated in HNSCC patients with miR-99a-3phigh miR-411-5plow miR-4746-5phigh expression
Selected genes related to invasion were compared, based on the significant relationship between 3 miRNA clusters with PNI status. As shown in Figure 4C,D, invasion-related genes (matrix metallopeptidase family members MMP1, MMP2, MMP3, MMP7, MMP9, MMP11, MMP12, MMP13, MMP16, and MMP28, integrin subunit beta 3 ITGB3, secreted protein acidic and cysteine rich SPARC) were significantly downregulated in HNSCC samples with miR-99a-3phighmiR-411-5plowmiR-4746-5phigh expression.
Discussion
High risk HPV are associated with precancerous lesions and carcinomas of cervical, vulvar, vaginal, anal, penile and HNSCC (18,19). HPV16 is the most common type in HNSCC and results in a distinct HNSCC subtype with unique pathological, clinical, molecular and prognostic characteristics (2,7,18,20).
Several miRNAs, including miR-21, miR-143, miR-9, miR-145, and miR-199a were identified in regulating the expression of host and viral factors that are particularly responsible for viral transcription which leads to HPV-related cervical carcinogenesis (21). HPV viral E6/E7 oncogenes may contribute to the deregulation of cellular miRNA network and subsequently affect tumor progression (22). miRNA deregulation, including miR-9, miR-21, miR-29a, miR-100 and miR-106b, were also reported in HPV16+ HNSCC (23-25). miR-15a/miR-16, miR-143/miR-145 and the miR-106/miR-363 cluster were suggested to target common HPV mediated pathways such as E6/p53 and E7/pRb in HPV-induced carcinogenesis (14). Furthermore, high risk E6 and E7 proteins could deregulate miRNA expression by directly interacting with other regulatory cellular proteins and DNA methylation (26,27). However, the interaction between HPV16 and the cellular miRNA network, along with the molecular mechanism behind tumor progression were not clear.
By comparing miRNA expression with the clinicopathological parameters of HNSCC patients, we revealed that less PNI events were identified in HNSCC patients with miR-99a-3phigh miR-411-5plow miR-4746-5phigh. PNI is well established as an independent prognostic indicator for HNSCC patients (28,29). In addition, HPV+ HNSCC patients were less likely to develop PNI than HPV- HNSCC patients (30). We proposed that miR-99a-3p, miR-411-5p and miR-4746-5p may participate in HPV16+ HNSCC progression and influence the patients’ prognosis.
To further determine the potential function of miR-99a-3p, miR-411-5p and miR-4746-5p, we explored the predicted target genes of 3 DEMs from the miRDB database and performed functional enrichment analysis. Based on the GO analysis, the predicted target genes of the 3 DEMs were enriched in cell growth, neuroepithelial cell differentiation, neuron projection extension, and neuron migration. KEGG pathway analysis showed that the predicted target genes were enriched in the MAPK signaling pathway and FoxO signaling pathway. The MAPK pathway is essential for the regulation of many cellular processes, including inflammation, cell proliferation, motility and apoptosis (31-33). It is well established that this pathway participates in HNSCC progression. Loss of FoxO function leads to increased cellular survival and plays a role in epithelial cancer progression (34).
We further screened DEGs in HNSCC patients with miR-99a-3phigh miR-411-5plow miR-4746-5phigh expression. GSEA revealed genes related to the EMT pathway were significantly downregulated in HNSCC patients with miR-99a-3phigh miR-411-5plow miR-4746-5phigh expression. Additionally, genes related to invasion were also downregulated, including matrix metallopeptidase family members MMP1, MMP2, MMP3, MMP7, MMP9, MMP11, MMP12, MMP13, MMP16, and MMP28, integrin subunit beta 3 ITGB3, secreted protein acidic and cysteine rich SPARC (35-37). Overexpression of EMT related genes were widely reported in the perineural invasion (PNI) sites of many cancers, including pancreatic cancer, colorectal adenocarcinoma, salivary adenoid cystic carcinoma and HNSCC (38-41).
MMPs, expressed in cancer or stromal cells, are capable of degrading ECM to promote tumor invasion and metastasis (35,42). The overexpression of MMPs, including MMP1, MMP3, MMP9, MMP11, and MMP13, is considered a risk factor for PNI in several cancers (43-46). Saygili et al. showed that MMP2 activation, under frequency electrical field stimulation, contributed to pro-NGF conversion and induced nerve sprouting (47). The overexpression of MMPs, such as MMP2 and MMP9, has also been widely reported in HNSCC tissues. However, the molecular mechanisms in PNI have not been widely investigated (48-51). The present study indicated potential roles of miR-99a-3p, miR-411-5p and miR-4746-5p in EMT and PNI of HPV16+ HNSCC.
HPV+ HNSCC patients have displayed significantly more favorable outcomes compared to HPV- HNSCC patients? (52,53). In addition, HNSCC patients with different HPV statuses exhibit distinct clinical presentations, socioeconomic, and molecular profiles (6,54,55). However, predictive biomarkers for the outcomes specific to HPV-related HNSCC patients are not clear. miRNAs, which act as posttranscriptional gene regulators, have been shown to be involved in almost all key cellular processes in cancer progression. Tissue-specific miRNAs have been explored as potential biomarkers in many kinds of cancers (56-59). A variety of prognostic miRNA biomarkers have also been selected for HNSCC, such as let-7d, miR-205, miR-99b-3p, and miR-100a-5p (60,61).
In this study, DEMs were screened in HPV16-related HNSCC samples, based on the TCGA database. Through subsequent multivariate Cox regression analysis, miR-99a-3p, miR-411-5p and miR-4746-5p were selected as potential prognostic biomarkers for HPV16(−) HNSCC (Figure 1). miR-99a-3p and miR-4746-5p were upregulated, and miR-411-5p was downregulated significantly in HNSCC tissues (Figure 2A). In addition, miR-99a-3p and miR-4746-5p predicted better outcomes, whereas miR-4746-5p forecasted poorer outcomes in HNSCC patients. As shown in Figure 4, patients with miR-99a-3plow miR-411-5phigh miR-4746-5plow expression exhibited poorer OS. The dysregulation of 3 DEMs in HPV16+ cancer samples could serve as a predictor of improved prognosis in HPV16+ HNSCC patients. Previous investigations have demonstrated that miR-99a-3p is downregulated in prostate cancer, colorectal cancer and HNSCC (62-65). Furthermore, Okada et al. observed significantly suppressed cell migration and invasion of FaDu and SAS cells, post miR-99a-3p transfection (62). This demonstrated the potential antitumor function of miR-99a-3p and highlighted its potential as a predictive biomarker for the positive prognosis of HNSCC, consistent with the findings of this study. miR-411-5p is regarded as a tumor suppressor in non-small cell lung cancer, hepatocellular carcinoma, and breast cancer. However, its function in HNSCC has not yet been reported (66-68). Kumaran et al. explored breast tissue-specific expression of miR-4746-5p compared to adjacent normal tissue; however, no further study was conducted (69). In addition to miR-99a-3p, our study showed that miR-411-5p and miR-4746-5p could also serve as potential prognostic biomarkers for HPV16-related HNSCC.
Overall, we determined that miR-99a-3p, miR-411-5p and miR-4746-5p are expressed differently in HPV16+ HNSCC patients. HPV16+ patients showed less PNI and more favorable OS with miR-99a-3phighmiR-411-5plowmiR-4746-5phigh expression. Further functional analysis showed downregulation of EMT gene sets and invasion related genes in HNSCC patients with miR-99a-3phighmiR-411-5plowmiR-4746-5phigh expression. The above results indicated possible roles of miR-99a-3p, miR-411-5p and miR-4746-5p in HPV16+ HNSCC progression. Furthermore, more comprehensive studies are required to affirm the MFs of the 3 miRNAs in HPV+ HNSCC.
Acknowledgments
Funding: The authors gratefully acknowledge the support obtained for this research from the Jiangsu Province Science & Technology Department (BE2018618), Nanjing Department of Health (YKK18120), Nanjing Medical Science and technique Development Foundation (QRX17174), the Key Project of Natural Science Research for College and University of Anhui Province (KJ2019A0356).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-6338
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-6338). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bravo IG, de Sanjosé S, Gottschling M. The clinical importance of understanding the evolution of papillomaviruses. Trends Microbiol 2010;18:432-8. [Crossref] [PubMed]
- Cullen M, Boland JF, Schiffman M, et al. Deep sequencing of HPV16 genomes: A new high-throughput tool for exploring the carcinogenicity and natural history of HPV16 infection. Papillomavirus Res 2015;1:3-11. [Crossref] [PubMed]
- Lowy DR. HPV vaccination to prevent cervical cancer and other HPV-associated disease: from basic science to effective interventions. J Clin Invest 2016;126:5-11. [Crossref] [PubMed]
- Koneva LA, Zhang Y, Virani S, et al. HPV Integration in HNSCC Correlates with Survival Outcomes, Immune Response Signatures, and Candidate Drivers. Mol Cancer Res 2018;16:90-102. [Crossref] [PubMed]
- Wagner S, Reder H, Sharma SJ, et al. HPV-associated oropharyngeal cancer-incidence, trends, diagnosis, and treatment. Urologe A 2018;57:1457-63. [Crossref] [PubMed]
- Zhu Y, Xia X, Gao L, et al. Prognostic implications of human papillomavirus type 16 status in non-oropharyngeal head and neck cancer: a propensity score matching analysis. Ann Transl Med 2019;7:759. [Crossref] [PubMed]
- Sano D, Oridate N. Pretreatment prognostic factor for patients with human papillomavirus related oropharyngeal cancer. Transl Cancer Res 2019;8:354-6. [Crossref]
- Satapathy S, Batra J, Jeet V, et al. MicroRNAs in HPV associated cancers: small players with big consequences. Expert Rev Mol Diagn 2017;17:711-22. [Crossref] [PubMed]
- Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer 2015;15:321-33. [Crossref] [PubMed]
- Zheng ZM, Wang X. Regulation of cellular miRNA expression by human papillomaviruses. Biochim Biophys Acta 2011;1809:668-77. [Crossref] [PubMed]
- Park S, Eom K, Kim J, et al. MiR-9, miR-21, and miR-155 as potential biomarkers for HPV positive and negative cervical cancer. BMC Cancer 2017;17:658. [Crossref] [PubMed]
- Sannigrahi MK, Sharma R, Singh V, et al. Role of Host miRNA Hsa-miR-139-3p in HPV-16-Induced Carcinomas. Clin Cancer Res 2017;23:3884-95. [Crossref] [PubMed]
- Emmett S, Whiteman DC, Panizza BJ, et al. An Update on Cellular MicroRNA Expression in Human Papillomavirus-Associated Head and Neck Squamous Cell Carcinoma. Oncology 2018;95:193-201. [Crossref] [PubMed]
- Lajer CB, Garnæs E, Friis-Hansen L, et al. The role of miRNAs in human papilloma virus (HPV)-associated cancers: bridging between HPV-related head and neck cancer and cervical cancer. Br J Cancer 2012;106:1526-34. [Crossref] [PubMed]
- Botezatu A, Goia-Rusanu CD, Iancu IV, et al. Quantitative analysis of the relationship between microRNA-124a, -34b and -203 gene methylation and cervical oncogenesis. Mol Med Rep 2011;4:121-8. [PubMed]
- Bardou P, Mariette J, Escudié F, et al. jvenn: an interactive Venn diagram viewer. BMC Bioinformatics 2014;15:293. [Crossref] [PubMed]
- Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 2015;43:D146-52. [Crossref] [PubMed]
- Araldi RP, Sant'Ana TA, Módolo DG, et al. The human papillomavirus (HPV)-related cancer biology: An overview. Biomed Pharmacother 2018;106:1537-56. [Crossref] [PubMed]
- Taberna M, Mena M, Pavón MA, et al. Human papillomavirus-related oropharyngeal cancer. Ann Oncol 2017;28:2386-98. [Crossref] [PubMed]
- Agalliu I, Gapstur S, Chen Z, et al. Associations of Oral α-, β-, and γ-Human Papillomavirus Types With Risk of Incident Head and Neck Cancer. JAMA Oncol 2016;2:599-606. [Crossref] [PubMed]
- Shishodia G, Verma G, Das BC, et al. miRNA as viral transcription tuners in HPV-mediated cervical carcinogenesis. Front Biosci (Schol Ed) 2018;10:21-47. [Crossref] [PubMed]
- Honegger A, Schilling D, Sültmann H, et al. Identification of E6/E7-Dependent MicroRNAs in HPV-Positive Cancer Cells. Methods Mol Biol 2018;1699:119-34. [Crossref] [PubMed]
- Sannigrahi MK, Sharma R, Singh V, et al. DNA methylation regulated microRNAs in HPV-16-induced head and neck squamous cell carcinoma (HNSCC). Mol Cell Biochem 2018;448:321-33. [Crossref] [PubMed]
- Božinović K, Sabol I, Dediol E, et al. Genome-wide miRNA profiling reinforces the importance of miR-9 in human papillomavirus associated oral and oropharyngeal head and neck cancer. Sci Rep 2019;9:2306. [Crossref] [PubMed]
- Sannigrahi MK, Sharma R, Panda NK, et al. Role of non-coding RNAs in head and neck squamous cell carcinoma: A narrative review. Oral Dis 2018;24:1417-27. [Crossref] [PubMed]
- McMurray HR, McCance DJ. Human papillomavirus type 16 E6 activates TERT gene transcription through induction of c-Myc and release of USF-mediated repression. J Virol 2003;77:9852-61. [Crossref] [PubMed]
- Wiklund ED, Gao S, Hulf T, et al. MicroRNA alterations and associated aberrant DNA methylation patterns across multiple sample types in oral squamous cell carcinoma. PLoS One 2011;6:e27840. [Crossref] [PubMed]
- Schmitd LB, Scanlon CS, D'Silva NJ. Perineural Invasion in Head and Neck Cancer. J Dent Res 2018;97:742-50. [Crossref] [PubMed]
- Liebig C, Ayala G, Wilks JA, et al. Perineural Invasion in Cancer A Review of the Literature. Cancer 2009;115:3379-91. [Crossref] [PubMed]
- Zhang Z, Liu R, Jin R, et al. Integrating Clinical and Genetic Analysis of Perineural Invasion in Head and Neck Squamous Cell Carcinoma. Front Oncol 2019;9:434. [Crossref] [PubMed]
- Chan L-P, Liu C, Chiang F-Y, et al. IL-8 promotes inflammatory mediators and stimulates activation of p38 MAPK/ERK-NF-kappa B pathway and reduction of JNK in HNSCC. Oncotarget 2017;8:56375-88. [Crossref] [PubMed]
- Lakshmanachetty S, Balaiya V, High WA, et al. Loss of TP63 Promotes the Metastasis of Head and Neck Squamous Cell Carcinoma by Activating MAPK and STAT3 Signaling. Mol Cancer Res 2019;17:1279-93. [Crossref] [PubMed]
- Leelahavanichkul K, Amornphimoltham P, Molinolo AA, et al. A role for p38 MAPK in head and neck cancer cell growth and tumor-induced angiogenesis and lymphangiogenesis. Molecular Oncology 2014;8:105-18. [Crossref] [PubMed]
- Reagan-Shaw S, Ahmad N. The role of Forkhead-box Class O (FoxO) transcription factors in cancer: a target for the management of cancer. Toxicol Appl Pharmacol 2007;224:360-8. [Crossref] [PubMed]
- Huang H. Matrix Metalloproteinase-9 (MMP-9) as a Cancer Biomarker and MMP-9 Biosensors: Recent Advances. Sensors (Basel) 2018;18:3249. [Crossref] [PubMed]
- Broek RV, Mohan S, Eytan DF, et al. The PI3K/Akt/mTOR axis in head and neck cancer: functions, aberrations, cross-talk, and therapies. Oral Diseases 2015;21:815-25. [Crossref] [PubMed]
- Chin D, Boyle GM, Williams RM, et al. Novel markers for poor prognosis in head and neck cancer. Int J Cancer 2005;113:789-97. [Crossref] [PubMed]
- Fujii-Nishimura Y, Yamazaki K, Masugi Y, et al. Mesenchymal-epithelial transition of pancreatic cancer cells at perineural invasion sites is induced by Schwann cells. Pathol Int 2018;68:214-23. [Crossref] [PubMed]
- Lin C, Ren Z, Yang X, et al. Nerve growth factor (NGF)-TrkA axis in head and neck squamous cell carcinoma triggers EMT and confers resistance to the EGFR inhibitor erlotinib. Cancer Lett 2020;472:81-96. [Crossref] [PubMed]
- Al Khatib AM, Mărgăritescu C, Taisescu O, et al. Immunoexpression of E-cadherin, Snail and Twist in colonic adenocarcinomas. Rom J Morphol Embryol 2019;60:531-6. [PubMed]
- Yang X, Jing D, Liu L, et al. Downregulation of p53 promotes in vitro perineural invasive activity of human salivary adenoid cystic carcinoma cells through epithelial-mesenchymal transition-like changes. Oncol Rep 2015;33:1650-6. [Crossref] [PubMed]
- Merchant N, Nagaraju GP, Rajitha B, et al. Matrix metalloproteinases: their functional role in lung cancer. Carcinogenesis 2017;38:766-80. [Crossref] [PubMed]
- Zhai LL, Wu Y, Cai CY, et al. Upregulated matrix metalloproteinase-2 and downregulated tissue factor pathway inhibitor-2 are risk factors for lymph node metastasis and perineural invasion in pancreatic carcinoma. Onco Targets Ther 2015;8:2827-34. [Crossref] [PubMed]
- Foda AAM, Aziz AA, Mohamed MA. Colorectal signet ring cell carcinoma: Influence of EGFR, E-cadherin and MMP-13 expression on clinicopathological features and prognosis. Ann Diagn Pathol 2018;32:41-6. [Crossref] [PubMed]
- Islekel H, Oktay G, Terzi C, et al. Matrix metalloproteinase-9,-3 and tissue inhibitor of matrix metalloproteinase-1 in colorectal cancer: relationship to clinicopathological variables. Cell Biochem Funct 2007;25:433-41. [Crossref] [PubMed]
- Li WM, Wei YC, Huang CN, et al. Matrix metalloproteinase-11 as a marker of metastasis and predictor of poor survival in urothelial carcinomas. J Surg Oncol 2016;113:700-7. [Crossref] [PubMed]
- Saygili E, Schauerte P, Pekassa M, et al. Sympathetic neurons express and secrete MMP-2 and MT1-MMP to control nerve sprouting via pro-NGF conversion. Cell Mol Neurobiol 2011;31:17-25. [Crossref] [PubMed]
- Stanciu AE, Zamfir-Chiru-Anton A, Stanciu MM, et al. Imbalance between Matrix Metalloproteinases and Tissue Inhibitors of Metalloproteinases Promotes Invasion and Metastasis of Head and Neck Squamous Cell Carcinoma. Clin Lab 2017;63:1613-20. [Crossref] [PubMed]
- Stanciu AE, Anton AZC, Stanciu MM, et al. Serum matrix metalloproteinase-2 in head and neck squamous cell carcinoma is associated with tumor differentiation. Romanian Biotechnological Letters 2017;22:12419-26.
- Stanciu AE, Zamfir-Chiru-Anton A, Stanciu MM, et al. Serum Level of Matrix Metalloproteinase-9 in Patients with Head and Neck Squamous Cell Carcinoma. Clin Lab 2016;62:1569-74. [Crossref] [PubMed]
- Kalfert D, Ludvikova M, Topolcan O, et al. Analysis of Preoperative Serum Levels of MMP1, -2, and -9 in Patients with Site-specific Head and Neck Squamous Cell Cancer. Anticancer Res 2014;34:7431-41. [PubMed]
- Fakhry C, Westra WH, Wang SJ, et al. The prognostic role of sex, race, and human papillomavirus in oropharyngeal and nonoropharyngeal head and neck squamous cell cancer. Cancer 2017;123:1566-75. [Crossref] [PubMed]
- Ang KK, Harris J, Wheeler R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med 2010;363:24-35. [Crossref] [PubMed]
- Braakhuis BJ, Snijders PJ, Keune WJ, et al. Genetic patterns in head and neck cancers that contain or lack transcriptionally active human papillomavirus. J Natl Cancer Inst 2004;96:998-1006. [Crossref] [PubMed]
- Smeets SJ, Braakhuis BJ, Abbas S, et al. Genome-wide DNA copy number alterations in head and neck squamous cell carcinomas with or without oncogene-expressing human papillomavirus. Oncogene 2006;25:2558-64. [Crossref] [PubMed]
- Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol 2009;4:199-227. [Crossref] [PubMed]
- Mirzaei H, Gholamin S, Shahidsales S, et al. MicroRNAs as potential diagnostic and prognostic biomarkers in melanoma. Eur J Cancer 2016;53:25-32. [Crossref] [PubMed]
- Moridikia A, Mirzaei H, Sahebkar A, et al. MicroRNAs: Potential candidates for diagnosis and treatment of colorectal cancer. J Cell Physiol 2018;233:901-13. [Crossref] [PubMed]
- Nassar FJ, Nasr R, Talhouk R. MicroRNAs as biomarkers for early breast cancer diagnosis, prognosis and therapy prediction. Pharmacol Ther 2017;172:34-49. [Crossref] [PubMed]
- Childs G, Fazzari M, Kung G, et al. Low-level expression of microRNAs let-7d and miR-205 are prognostic markers of head and neck squamous cell carcinoma. Am J Pathol 2009;174:736-45. [Crossref] [PubMed]
- Jakob M, Mattes LM, Kuffer S, et al. MicroRNA expression patterns in oral squamous cell carcinoma: hsa-mir-99b-3p and hsa-mir-100-5p as novel prognostic markers for oral cancer. Head Neck 2019;41:3499-515. [Crossref] [PubMed]
- Okada R, Koshizuka K, Yamada Y, et al. Regulation of Oncogenic Targets by miR-99a-3p (Passenger Strand of miR-99a-Duplex) in Head and Neck Squamous Cell Carcinoma. Cells 2019;8:1535. [Crossref] [PubMed]
- Wu C, Tong L, Wu C, et al. Two miRNA prognostic signatures of head and neck squamous cell carcinoma: A bioinformatic analysis based on the TCGA dataset. Cancer Med 2020;9:2631-42. [Crossref] [PubMed]
- Wei GG, Guo WP, Tang ZY, et al. Expression level and prospective mechanism of miRNA-99a-3p in head and neck squamous cell carcinoma based on miRNA-chip and miRNA-sequencing data in 1, 167 cases. Pathol Res Pract 2019;215:963-76. [Crossref] [PubMed]
- Arai T, Okato A, Yamada Y, et al. Regulation of NCAPG by miR-99a-3p (passenger strand) inhibits cancer cell aggressiveness and is involved in CRPC. Cancer Med 2018;7:1988-2002. [Crossref] [PubMed]
- Zhang C, Wang H, Liu X, et al. Oncogenic microRNA-411 promotes lung carcinogenesis by directly targeting suppressor genes SPRY4 and TXNIP. Oncogene 2019;38:1892-904. [Crossref] [PubMed]
- Zeng Z, Li Y, Pan Y, et al. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat Commun 2018;9:5395. [Crossref] [PubMed]
- Zhang Y, Xu G, Liu G, et al. miR-411-5p inhibits proliferation and metastasis of breast cancer cell via targeting GRB2. Biochem Biophys Res Commun 2016;476:607-13. [Crossref] [PubMed]
- Kumaran M, Krishnan P, Cass CE, et al. Breast cancer associated germline structural variants harboring small noncoding RNAs impact post-transcriptional gene regulation. Sci Rep 2018;8:7529. [Crossref] [PubMed]
(English Language Editor: E. Tan)


