
Multi-omics analysis reveals the genetics and immune landscape of dexamethasone responsive genes in cancer microenvironment
Introduction
Glucocorticoids are widely used in the treatment of cancer (1,2). It is widely accepted that using glucocorticoids can relieve symptoms of cancer and its treatment in the clinical practice (1,2). For example, glucocorticoids prevent vomiting and allergic reactions associated with cancer therapy (2,3). Glucocorticoids decrease edema in CNS malignancy (3), and can decrease pain secondary to cancer (4). As the adjuvant antitumor therapy glucocorticoids can relieve superior vena cava syndrome, increased intracranial pressure, spinal cord compression or autoimmune disease caused by tumor (5). On the contrary, glucocorticoids can prevent tumor-related adverse events (6), which are mainly used for pretreatment before the administration of certain anti-tumor drugs, antiemetic treatment of chemo-related to radiotherapy and chemotherapy, and anti-inflammatory treatment of inflammatory damage (7,8).
However, in the context of immunotherapy, two independent groups report that using Glucocorticoids can negatively impact the patient outcome during the course of immune checkpoint inhibition (9,10). For example, in advanced melanoma patients, baseline corticosteroid use contributes to poorer overall survival (HR 1.23) (9,10). These observations suggest that avoiding corticosteroid or combining other agents should be considered at the initiation of immune checkpoint inhibition treatment, although the underlying mechanism still remains unclear (11). We hence hypothesize that the glucocorticoids usage may reprogram the tumor microenvironment and hence impacts the immunotherapy efficacy.
The cancer microenvironment is composed not only of a heterogeneous population of tumor cells but also a number of infiltrating immune cells (12). Its constitution determines the cancer cell fate and clinical phenotype. Under such a scenario, understanding drug-microenvironment interaction is fundamental for designing combinational agent and patient treatment strategy. For example, targeting the hypoxia microenvironment can boost the efficacy of PD-L1 immunotherapy (13). Therefore, there is a pressing need to explore how dexamethasone associates the cancer microenvironment and how dexamethasone responsive genes correlate with infiltrated immune cells. Given the tumor microenvironment is a complex ecosystem which not only consists of cancer cells but also immune cells, we hypothesize that dexamethasone usage will also impact the infiltrated T cells.
Here, by using pan-cancer multi-omics data of thousands of samples, we seek to define the landscape of dexamethasone responsive genes and how they associate with cancer hallmarks. We comprehensively analyze signaling pathways dexamethasone responsive pathways and observe that dexamethasone tightly associates with cancer microenvironment. Our study provides novel insights linking dexamethasone response with infiltrating immune cells and cancer microenvironment.
Methods
Data source and data availability
We fetched the RNA-seq data from The Cancer Genome Atlas (http://api.gdc.cancer.gov/data). Copy number variation (CNV) data was downloaded from UCSC Xena (https://gdc.xenahubs.net/download/GDC-PANCAN.gistic.tsv.gz). Methylation data was downloaded from UCSC Xena (https://gdc.xenahubs.net/download/GDC-PANCAN.methylation450.tsv.gz). Survival data was downloaded from UCSC Xena (https://gdc.xenahubs.net/download/GDC-PANCAN.survival.tsv.gz). SNP data was downloaded from UCSC Xena (https://gdc.xenahubs.net/download/GDC-PANCAN.mutect2_snv.tsv.gz). All raw data are available by request. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Transcriptome analysis and prognosis analysis
We analyzed RNA-seq data cancers with matched normal samples in The Cancer Genome Atlas (THCA, KIRP, LIHC, STAD, BRCA, COAD, UCEC, BLCA, KIRC, KICH, and PRAD). We performed the differential expression analysis based on the count data by using DESeq2 (14) as is described before (15,16). Genes with fold change over two and P value less than 0.05 were considered as the significantly differentially expressed genes. As for the correlation analysis, we used the spearman test of normalized expression data by using R. To compute the dexamethasone response score, we used ssGSEA algorithm based on dexamethasone responsive genes in all primary cancer samples (17).
CNV analysis and single nucleotide variation (SNV) analysis
We used GISTIC to process the CNV data and annotate the CNV type (18). All CNV events were divided in heterozygous amplification and heterozygous deletion. Only events with over 5% heterozygous amplification or deletion were included for analysis. To compute the correlation CNV and RNA expression, we used cor.test function in R. As for SNV analysis, we used maftools to annotate raw SNV data (19). All SNV events were classified into splice site, frameshift insertion, in-frame deletion, nonstop mutation, mis sense mutation, non-sense mutation, and frame shift deletion. We ranked all SNV events according to the variant type, variant classification, SNV class, variants per sample, and gene mutation frequency.
Signalling pathway analysis
We first divided all samples into the higher expression group and lower expression based on the expression value of a given gene. We next computed the pathway score of ten hallmark signalling pathways (TSC/mTOR, RTK, RAS/MAPK, PI3K/AKT, Hormone estrogen receptor pathway, Hormone androgen receptor pathway, Epithelial–mesenchymal transition, DNA Damage Response, Cell Cycle, and Apoptosis pathway) based on the reverse phase protein array value from The Cancer Proteome Atlas (http://tcpaportal.org/tcpa/). We next compared the pathway score based on the patient group defined by the previous analysis.
Tumor microenvironment analysis
We downloaded the tumor microenvironment cell (immune cell or stroma cell) score matrix in xCell (20). As for the correlation analysis, we used the cor.test function in R.
Drug response correlation analysis
We fetched the frug response profile (AUC value) and gene expression profile from Therapeutics Response Portal (https://portals.broadinstitute.org/ctrp/) (21). We next performed the Pearson correlation analysis between RNA expression and drug response value by using cor.test in R.
Statistical analysis
P value or FDR less than 0.05 was regarded as statistical significance. For the differential expression analysis, fold change >2 and P value <0.05 was considered as statistical significance. As for the correlation analysis, spearman rho over 0.3 and P value less than 0.05 was regarded as statistical significance.
Results
Dexamethasone responsive genes are generally downregulated and are associated with worse patient survival
In order to get a systematic understanding of dexamethasone responsive genes, we extracted the gene sets from Gene Ontology (cellular response to dexamethasone stimulus) including 25 genes (AGTR1, AGTR2, AQP1, BMP4, CASP9, DDIT4, EIF4E, EIF4EBP1, FBXO32, GDNF, HNRNPU, IFNB1, JAK2, MSTN, NR3C1, PCK1, PCK2, RPS6KB1, SMYD3, TFAP4, TGFB1, TRIM63, C13orf39, IFNA17, IFNA2). We next downloaded the transcriptome and matched phenotype data from The Cancer Genome Atlas. We interestingly observed that more dexamethasone responsive genes are downregulated in a wide range of cancers, while a small fraction of them are downregulated (n=716, Figure 1A). Among all significant alternations, 61 of them are downregulated and 35 are upregulated. This observation suggested that tumors are likely to be less responsive to dexamethasone compared with normal cells. NR3C1 is the receptor of dexamethasone (22). The expression of such gene may also impact the dexamethasone response of tumors. We hence analyze the differential expression profile of NR3C1 (Figure S1A). We also analyze the methylation profile and expression profile in different cancer subtypes (Figure S1B,C,D). Next, we tested whether the aberrant dexamethasone responsive gene expression is correlated with overall survival. To our surprise, patients with high expression of dexamethasone responsive genes showed higher survival risk (Figure 1B). Next, we asked the dexamethasone responsive landscape across different cancers. We used the ssGSEA algorithm to compute the enrichment score of dexamethasone responsive genes. As a result, dexamethasone response score was higher in cancers such as liver hepatocellular carcinoma (LIHC) or skin cutaneous melanoma (SKCM). Testicular germ cell tumors showed the lowest dexamethasone response score, indicating that dexamethasone responsive genes are dysregulated in this cancer (Figure 1C). Collectively, our finding demonstrated that cancer cells widely downregulated dexamethasone responsive genes and may be less responsive to dexamethasone.
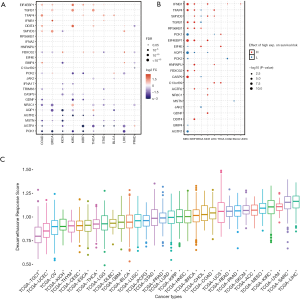
Dexamethasone responsive genes show high frequency of heterozygous deletion in cancers
To explore why the dexamethasone responsive genes are downregulated in cancers, we performed copy number variation (CNV) analysis across 11 cancer types. In general, CNV of dexamethasone responsive genes was in consistence with their RNA expression (Figure 2A). For example, the upregulated genes, such as TFAP4 and EIF4EBP1, were significantly heterozygous amplified in 10 cancers such as breast cancer and liver cancer (Figure 2A, Figure S2). On the contrary, AGTR2, whose transcriptional expression was downregulated is 5 cancers, was frequently heterozygous deleted (Figure 2A). We next analyzed the homozygous amplification and deletion of those genes and observed that such events are rare across cancers (Figure 2B). Those observation suggested that the dysregulation dexamethasone responsive possibly arises from the CNV. To validate our result, we checked the correlation between RNA expression and CNV profile. As expected, the majority of genes showed high correlation between CNV and transcript expression (Figure 2C). However, CNV was not always correlated with expression profile. For example, PCK1 and AQP1 expression were transcriptionally downregulated, but they showed heterozygous amplification in most cancers. In summary, heterozygous deletion of dexamethasone responsive genes may be predominate in pan-cancer which likely contributes to the global transcriptional downregulation.
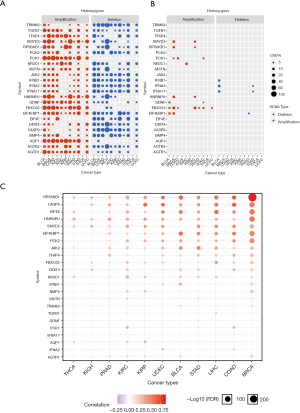
Dexamethasone responsive genes are frequently mutated in cancers
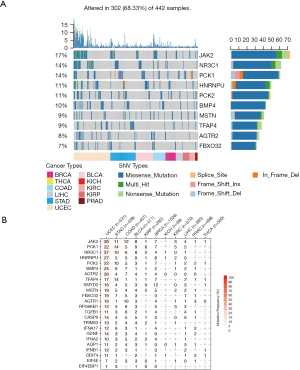
SNV is a single nucleotide mutation that occurs at a specific position in the genome, where each variation is at a level of >1% in the population. SNV widely exists and may impacts the protein function. We hence downloaded the SNV profile (Methods) of those 25 genes. To our surprise, in certain cancers, dexamethasone responsive genes were highly mutated (Figure 3A, n=442). For example, in uterine corpus endometrial carcinoma, 39% samples showed JAK2 mutation (Figure 3B). In pan-cancer, JAK2 is also frequently mutated (17% samples). Among all mutation events, missense mutations were most predominant where C>T and C>A are the top SNV events (Figure S3A,B,C). However, not all mutated genes showed transcriptional dysregulation. For example, JAK2 that was mutated in 7% breast cancer samples, however, it did not have a difference in RNA expression in breast cancer samples and normal samples. In summary, dexamethasone responsive genes showed high single nucleotide mutation profile and this may explain their transcriptional dysregulation in cancers.
Dexamethasone responsive genes correlate with cancer hallmarks
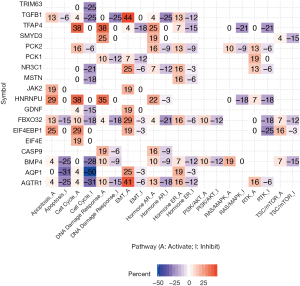
We next hypothesized dexamethasone response may associate with cancer hallmark pathways. To test this hypothesis, we selected 10 hallmark pathways and compare their enrichment score based on gene expression (Figure 4, Figure S4). To our surprise, dexamethasone responsive gene related pathways were enriched in epithelial-to-mesenchymal transformation (EMT) activation and cell cycle inhibition (Figure 4). For example, AQP1 was related with inhibition of cell cycle gene sets in 50% samples. AGTR1, AQP1, EIF4EBP1, FBXO32, GDNF, JAK2, NR3C1, PCK1, TGFB1, and TFAP4 associated with EMT activation (Figure 4). To sum up, dexamethasone response was potentially enriched in epithelial-to-mesenchymal transformation and cell cycle inhibition.
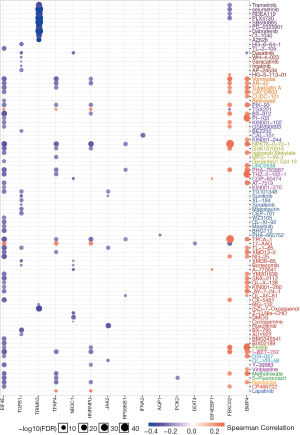
Dexamethasone responsive gene expression tightly associates with sensitivity of 90 anti-cancer drug
To explore potential combinational therapy with dexamethasone, a crucial step is to evaluate the association between dexamethasone response genes and anti-cancer agent effectiveness profile. To compute the association of dexamethasone response gene and drug efficacy, the spearman rho of RNA expression and AUCs was used and normalized based on Z-score from CTRP (Methods). As a result, BMP4 and FBXO32 showed positive correlation with a large amount of drugs such as Methotrexate and Navitoclax (Figure 5). On the contrary, EIF4E, TGFB1, TRIM63, TFAP4, NR3C1, HNRNPU, JAK2, and RPS6KB1 showed negative correlation with existed anticancer drugs (Figure 5). In a nutshell, we screened the potentially compatible drugs of dexamethasone and provided a potential combinational drug list which may be useful for the oncology community.
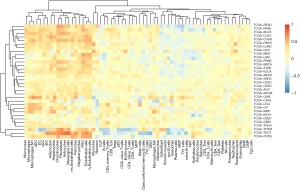
Dexamethasone response positively associates with the innate immune response
Given that our previous observation that dexamethasone responsive genes are enriched in several immune-related pathways such as EMT, we questioned whether dexamethasone response may associate with tumor-infiltrated immune cells. We used the ssGSEA algorithm to compute the enrichment score of dexamethasone responsive genes. We next evaluated the correlation between dexamethasone response score and CD8 T cells, CD4 T cells, B cells, dendritic cells, macrophages, neutrophils, etc. As expected, dexamethasone response score negatively correlated with almost half of immune cells inside the tumor microenvironment (Figure 6, Figure S5). Interestingly, those immune cells were enriched in cellular immune cells (Figure 6). This may be due to the immune inhibitory role of dexamethasone. On the contrary, we observed that a subset of immune cells positively associate with dexamethasone response (Figure 6). For example, in 20 cancer types such as melanoma, dexamethasone response associated with macrophage infiltration level. This trend was conserved in other innate immune cells including DCs and monocytes. In summary, our data indicated that dexamethasone response may promotes the innate immunity inside the tumor microenvironment, which provides further explanation that how dexamethasone exerts their anti-tumor effects.
Discussion
Although dexamethasone is widely used in cancer treatment, it is still unclear how dexamethasone response associate with cancer microenvironment and cancer hallmarks. In this paper, we attempt to define the landscape of dexamethasone responsive genes by utilizing multi-omics data in pan-cancer. We comprehensively analyze signaling pathways potentially linked with dexamethasone response and demonstrate that dexamethasone tightly associates with cancer microenvironment. We also provide a list of drug combination with dexamethasone based on drug sensitivity analysis. Our research provides a systematic resource of linking dexamethasone response with infiltrating immune cells and cancer microenvironment.
First, we demonstrate how dexamethasone responsive genes is transcriptional dysregulated. We integrate transcriptome data, methylation data, and CNV data from thousands of samples in pan-cancer. In the transcriptome level, dexamethasone responsive genes are generally downregulated. This indicates that cancer cells may not respond to dexamethasone like normal cells. We next demonstrate that, such downregulation originates from CNV of dexamethasone responsive genes. Those genes are heterozygous deleted across a broad spectrum of cancers. Our data, for the first time, report the dexamethasone responsive gene dysregulation across cancers.
Second, we provide a list of potential combinational drug pairs with dexamethasone. Glucocorticoids, such as dexamethasone, are widely used for prevent vomiting and allergic reactions associated with cancer therapy (23). However, the compatibility of dexamethasone has not been investigated in a systems level. We analyze the correlation between 367 compounds and dexamethasone responsive genes in 987 Cell lines. Our data indicate that, 90 drugs significantly associate with at least one dexamethasone-related gene. A subset of our result is in consistency with published results. For example, lapatinib, an epidermal growth factor receptor (EGFR) tyrosine kinases inhibitor, was previous reported to interact with dexamethasone. Our data show that dexamethasone responsive genes, such as TFAP4 and HNRNPU, positively associates with lapatinib drug sensitivity (24). On the contrary, our data also provide novel dexamethasone compatibility candidates. TGX221, a PI3K inhibitor (25), was observed to positively associate with dexamethasone responsive genes. In summary, our analysis not only confirm the published results but also provide new potential combinational drug pairs with dexamethasone.
Last, our analysis indicates the link between dexamethasone response and cancer microenvironment. Although Glucocorticoids can inhibit the systemic immune responses (26), nevertheless, how Glucocorticoids impacts immune cells, particularly inside the tumor microenvironment, still remains unclear. Given the tumor microenvironment is a complex ecosystem which not only consists of cancer cells but also immune cells, we perform the correlation analysis between all dexamethasone responsive genes and 64 immune cells/stroma cells inside the microenvironment. As expected, cellular immune cells show the negative correlation with the dexamethasone response. However, the innate immune cells are positively correlated with dexamethasone response and this trend are highly conserved in cancers. Those observation suggest the polarizing effect of dexamethasone on immune cells inside the cancer microenvironment. Further experimental or clinical results are required to confirm our findings in the future.
Our analyses do have some disadvantages. First, dexamethasone may have non-genomic effects on cells, where tumor cells may respond to it without changes of gene expression. Our in silico analysis of transcriptional changes of dexamethasone responsive genes may partly represents the functional states of cancer cells. Second, in our differential expression analysis data, only cancer types with matched normal samples were included. Hence, hematological neoplasms were not included for analysis. Third, our analysis cannot identify whether dexamethasone responsive genes be turned on and whether the patients have received dexamethasone. However, the goal of this project is to check whether the dexamethasone responsive genes are altered in tumors and how they varied across different cancer types. The gene expression effects may be due to Cortisol, the main endogenous glucocorticoid in humans. Forth, not all analysis show consistent result. For example, CNV can only partly explain expression level changes. For example, PCK1 and AQP1 expression transcriptionally downregulated, but they show heterozygous amplification in most cancers. Not all mutated genes show transcriptional dysregulation. For example, JAK2 that is mutated in 7% breast cancer samples, however, it does not have a difference in RNA expression in breast cancer samples and normal samples.
In summary, our study demonstrates the dysregulation of dexamethasone responsive genes in multiple cancers and such downregulation originates from CNV. Our analysis provides a list of potential combinational drug pairs with dexamethasone and indicate that dexamethasone usage may promote the innate immunity inside the tumor microenvironment.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-3650). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Haywood A, Duc J, Good P, et al. Systemic corticosteroids for the management of cancer-related breathlessness (dyspnoea) in adults. Cochrane Database Syst Rev 2019;2:CD012704. [Crossref] [PubMed]
- Vayne-Bossert P, Haywood A, Good P, et al. Corticosteroids for adult patients with advanced cancer who have nausea and vomiting (not related to chemotherapy, radiotherapy, or surgery). Cochrane Database Syst Rev 2017;7:CD012002. [Crossref] [PubMed]
- Roth P, Happold C, Weller M. Corticosteroid use in neuro-oncology: an update. Neurooncol Pract 2015;2:6-12. [Crossref] [PubMed]
- Leppert W, Buss T. The role of corticosteroids in the treatment of pain in cancer patients. Curr Pain Headache Rep 2012;16:307-13. [Crossref] [PubMed]
- Strehl C, Ehlers L, Gaber T, et al. Glucocorticoids-All-Rounders Tackling the Versatile Players of the Immune System. Front Immunol 2019;10:1744. [Crossref] [PubMed]
- Montgomery B, Cheng HH, Drechsler J, et al. Glucocorticoids and prostate cancer treatment: friend or foe? Asian J Androl 2014;16:354-8. [Crossref] [PubMed]
- Raetz EA, Loh ML, Devidas M, et al. Impact of corticosteroid pretreatment in pediatric patients with newly diagnosed B-lymphoblastic leukemia: a report from the Children's Oncology Group. Haematologica 2019;104:e517-20. [Crossref] [PubMed]
- Ma Y, Yang H, Kroemer G. Endogenous and exogenous glucocorticoids abolish the efficacy of immune-dependent cancer therapies. Oncoimmunology 2019;9:1673635. [Crossref] [PubMed]
- Drakaki A, Luhn P, Wakelee H, et al. 470 Association of systemic corticosteroids with overall survival in patients receiving cancer immunotherapy for advanced melanoma, non-small cell lung cancer or urothelial cancer in routine clinical practice. Ann Oncol 2019;30:xi16-7. [Crossref]
- De Giglio A, Mezquita L, Auclin E, et al. 460 Impact of early introduction of steroid on immune-checkpoint inhibitors (ICI) in patients with advanced non-small cell lung cancer treated. Ann Oncol 2019;30:xi16-7. [Crossref]
- Kelly WJ, Gilbert MR. Glucocorticoids and immune checkpoint inhibitors in glioblastoma. J Neurooncol 2020. [Epub ahead of print]. [Crossref] [PubMed]
- Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med 2018;24:541-50. [Crossref] [PubMed]
- Jiang X, Wang J, Deng X, et al. Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol Cancer 2019;18:10. [Crossref] [PubMed]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [Crossref] [PubMed]
- Wu Y, Yang Y, Gu H, et al. Multi-omics analysis reveals the functional transcription and potential translation of enhancers. Int J Cancer 2020;147:2210-24. [Crossref] [PubMed]
- Wu Y, Wei J, Chen X, et al. Comprehensive transcriptome profiling in elderly cancer patients reveals aging-altered immune cells and immune checkpoints. Int J Cancer 2019;144:1657-63. [Crossref] [PubMed]
- Foroutan M, Bhuva DD, Lyu R, et al. Single sample scoring of molecular phenotypes. BMC Bioinformatics 2018;19:404. [Crossref] [PubMed]
- Mermel CH, Schumacher SE, Hill B, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 2011;12:R41. [Crossref] [PubMed]
- Mayakonda A, Lin DC, Assenov Y, et al. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res 2018;28:1747-56. [Crossref] [PubMed]
- Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol 2017;18:220. [Crossref] [PubMed]
- Rees MG, Seashore-Ludlow B, Cheah JH, et al. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat Chem Biol 2016;12:109-16. [Crossref] [PubMed]
- Liu B, Zhang TN, Knight JK, et al. The Glucocorticoid Receptor in Cardiovascular Health and Disease. Cells 2019;8:1227. [Crossref] [PubMed]
- Van Ryckeghem F. Corticosteroids, the oldest agent in the prevention of chemotherapy-induced nausea and vomiting: What about the guidelines? J Transl Int Med 2016;4:46-51. [Crossref] [PubMed]
- Teo YL, Saetaew M, Chanthawong S, et al. Effect of CYP3A4 inducer dexamethasone on hepatotoxicity of lapatinib: clinical and in vitro evidence. Breast Cancer Res Treat 2012;133:703-11. [Crossref] [PubMed]
- Feng C, Sun Y, Ding G, et al. PI3Kbeta inhibitor TGX221 selectively inhibits renal cell carcinoma cells with both VHL and SETD2 mutations and links multiple pathways. Sci Rep 2015;5:9465. [Crossref] [PubMed]
- Coutinho AE, Chapman KE. The anti-inflammatory and immunosuppressive effects of glucocorticoids, recent developments and mechanistic insights. Mol Cell Endocrinol 2011;335:2-13. [Crossref] [PubMed]


