
Repressing PDCD4 activates JNK/ABCG2 pathway to induce chemoresistance to fluorouracil in colorectal cancer cells
Introduction
Colorectal cancer (CRC) is the third most common malignant tumor worldwide, with increasing morbidity and mortality. Surgical resection, chemotherapy, radiotherapy, targeted therapy, and immunotherapy are the main treatments for CRC (1). Fluorouracil (5-FU), an analog of uracil with a fluorine atom at the C-5 position in place of hydrogen, has been widely used to treat various cancers since 1957. It is still one of the main chemotherapeutic agents in CRC treatment (2,3). 5-FU-based chemotherapy has been shown to improve the overall and disease-free survival of patients with resectable stage III CRC. However, the occurrence of 5-FU resistance during treatment is one of the main causes of poor prognosis in advanced CRC. Thus, an improved understanding of the mechanisms underlying 5-FU resistance in CRC will provide insights into overcoming resistance.
Members of the adenosine triphosphate (ATP)-binding cassette (ABC) transporter superfamily have been found to be associated with multidrug resistance (MDR) in various cancers, including breast cancer, gastric cancer, and CRC (4). Several studies have demonstrated that ABC transporter superfamily members are responsible for MDR through pumping drug or drug metabolites out of cells. ABCB5 and ABCC10 have been reported to play a key role in 5-FU resistance by regulating c-Myc (5) and FOXM1 (6), respectively. Furthermore, loss of ABCB4 has been found in 5-FU-resistant CRC cells (7), and ABCB1, ABCC1, and ABCC4 have been shown to correlate with 5-FU resistance. It has been reported that fluoropyrimidine metabolites are substrates of ABCC5 and ABCC11 in a variety of cancer cells (8-10), and that 5-FU is a substrate of ABCG2 in breast cancer cells (11). Additionally, in both gastric cancer cells and colon cancer cells, ABCG2 has been found to be overexpressed in cancer stem-like cells, and was shown to be responsible for 5-FU resistance (10,12). Moreover, the c-Jun NH2 terminal kinase (JNK) pathway has been reported to be related to the overexpression of ABCG2 in cancer stem-like cells (13), as well as ABCG2-induced resistance to hydroxycamptothecin (HCPT) in SW1116 cells (14). Nevertheless, whether the JNK/ABCG2 pathway mediates 5-FU resistance in CRC cells and the details of the possible mechanisms involved are still unknown.
We hypothesized that the JNK/ABCG2 pathway may induce acquired resistance to 5-FU in CRC cells, and investigated the upstream mechanisms of JNK/ABCG2 in CRC cells. Herein, we established acquired 5-FU-resistant HCT116 cells (HCT116/FUR), and a 5-FU-resistant cell-derived xenograft nude mouse model. The studies in CRC cells and HCT116/FUR cells showed that microRNA-21 (miR-21) targeted and inhibited programmed cell death 4 (PDCD4), which was involved in the regulation of the JNK/ABCG2 pathway. Our in vivo and in vitro results suggested that inhibition of the JNK pathway overcame 5-FU resistance. Overall, our work demonstrates the critical role of the PDCD4/JNK/ABCG2 axis regulated by miR-21 in acquired resistance to 5-FU in CRC cells. These insights also provide potential therapeutic implications for 5-FU-resistant CRC patients.
We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-4292).
Methods
Materials
Dulbecco’s Modified Eagle’s Medium (DMEM) high glucose culture medium was purchased from Hyclone (GE Healthcare Life Science, NJ, USA), and fetal bovine serum (FBS) was purchased from Biological Industry (Göttingen, Germany). Dimethyl sulfoxide (DMSO), 3-(4, 5-dimethyl thiazol-2-yl)-2, 5-diphenyl tetrazolium bromide (MTT), and bovine serum albumin (BSA) were purchased from Solarbio (Shanghai, China). SP600125 and 5-FU were purchased from Sigma-Aldrich (St Louis, MO, USA). Antibodies specific for c-Jun (#380397), phospho-c-Jun (Ser63) (#382955), phospho-JNK (Thr183/Tyr185) (#619878), and b-actin (#200068-6D7) were obtained from Zen-Bio (Chengdu, China). Antibodies specific for ABCG2 (#27286-1-AP), PDCD4 (Cat# 12587-1-AP), horseradish peroxidase (HRP)-conjugated AffiniPure Goat anti-rabbit immunoglobin G (IgG) (H+L) (#SA00001-2), HRP-conjugated AffiniPure Goat anti-mouse IgG (H+L) (#SA00001-1), and GAPDH (#60004-1-Ig) were purchased from Proteintech Group, Inc (Rosemont, IL, USA). JNK antibody (#CY5490) was purchased from Abways Technology (China). SP600125 and 5-FU were dissolved in DMSO.
Cell culture
The human CRC cell lines HCT116, RKO, RKO-KO (microRNA-21 knockout), and HCT116/FUR were cultured in high glucose DMEM supplemented with 10% FBS, 100 U/mL of penicillin, and 100 U/mL of streptomycin (Hyclone, GE Healthcare Life Science, NJ, USA) at 37 °C in a humidified atmosphere containing 5% CO2. 5-FU-resistant HCT116 cells were established by culturing in continuously increasing concentrations of 5-FU for at least 6 months. RKO and HCT116 cells were purchased from the American Type Culture Collection (ATCC), and RKO-KO cells were kindly provided by Professor Lin Zhang from the University of Pittsburgh.
Small interfering RNA (siRNA), small hairpin RNA (shRNA), and plasmids for PDCD4 and ABCG2
PDCD4 shRNA and a small hairpin non-target control (shNT) were designed according to the cDNA sequence on the Broad Institute Website, and the small interfering RNAs (siRNAs) were purchased from Gene Pharma (Shanghai, China). The shRNA was constructed into pLKO.1-TRC plasmid. FLAG-tagged PDCD4 and HA-tagged ABCG2 cDNA were constructed with the standard polymerase chain reaction (PCR) technique using Phanta HS Super-Fidelity DNA Polymerase (Vazyme, Nanjing, China) into pcDNA3.1 vector, named PDCD4-FLAG and ABCG2-HA, respectively. Cells were transfected with 4 µg of the plasmid, or 100 pmol of siRNA using PEI Transfection Reagent (Sigma-Aldrich, St Louis, MO, USA) according to the manufacturer’s instructions. The sequences of primers, siRNAs, and shRNAs were as follows:
PDCD4: BamHI-PDCD4-F: 5'GAT CGG ATC CAT GAC CAA ATA TCT GAT AAC-3'; EcoRI-PDCD4-R: 5'-AAT TGA ATT CGT AGC TCT CTG GTT TAA GAC-3'.
ABCG2: BamHI-ABCG2-F: 5'-GAT CGG ATC CAT GTC TTC CAG TAA TGT CG-3'; EcoRI-ABCG2-R: 5'-AAT TGA ATT CCC AAA TAT TCT TCG CCA GTA C-3'.
shPDCD4: F: 5'-CCG GCG CCC TTA GAA GTG GAT TAA CCT CGA GGT TAA TCC ACT TCT AAG GGC GTT TTT G-3'; R:5'-AAT TCA AAA ACG CCC TTA GAA GTG GAT TAA CCT CGA GGT TAA TCC ACT TCT AAG GGC G-3'.
siNT (si non-targeting RNA): sense: 5'-UUC UCC GAA CGU GUC ACG UTT-3'; antisense: 5'-ACG UGA CAC GUU CGG AGA ATT-3'.
siABCG2: sense: 5'-UGU CUU AGC UGC AAG GAA ATT-3'; antisense: 5'-UUU CCU UGC AGC UAA GAC ATT-3'.
Mimic-miR-21: 5'-UAG CUU AUC AGA CUG AUG UUG A-3'. miR-21 inhibitor: 5'-UCA ACA UCA GUC UGA UAA GCU A-3'.
Semi-quantitative reverse transcription PCR
The total RNA of treated cells was extracted using RNAiso Plus (TaKaRa, Tokyo, Japan) according to the manufacturer’s instructions, then cDNA synthesis was carried out using HiScriptTM Q-RT Super Mix for quantitative PCR (qPCR) (+gDNA wiper) (Vazyme, Nanjing, China). Real-time PCR was performed using SYBR Green Master Mix (Vazyme, Nanjing, China) on the Bio-Rad CFX96 (Bio-Rad, CA, USA). All experiments were performed in triplicate, and were normalized to the internal reference gene glycer-aldehyde-3-phosphate dehydtrogenase (gapdh) or TATA box binding protein (tbp). Relative mRNA expression was calculated using the 2(-ΔΔCT) method. Primers for qRT-PCR were as follows:
GAPDH: F: 5'-TGC ACC ACC AAC TGC TTA GC-3'; R: 5'-GGC ATG GAC TGT GGT CAT GAG-3'.
TBP: F: 5'-CCC ATG ACT CCC ATG ACC-3'; R: 5'-TTT ACA ACC AAG ATT CAC TGT GG-3'.
Mouse TBP: F: 5'-GTG AAG GGT ACA AGG GGG TG-3'; R: 5'-ACA TCT CAG CAA CCC ACA CA-3'.
PDCD4: F: 5'-GAT TAA CTG TGC CAA CCA GTC CAA AG-3'; R: 5'-CAT CCA CCT CCT CCA CAT CAT ACA C-3'.
ABCG2: F: 5'-CAG GTG GAG GCA AAT CTT CGT-3'; R: 5'-ACC CTG TTA ATC CGT TCG TTT T-3'.
Mouse ABCG2: F: 5'-GAA CTC CAG AGC CGT TAG GAC-3'; R: 5'-CAG AAT AGC ATT AAG GCC AGG TT-3'.
miR-21: miR-21-RT: 5'-GTC GTA TCC AGT GCA GGG TCC GAG GTA TTC GCA CTG GAT ACG ACT CAA CAT-3'.
miR-21: F: 5'-TGC CGC CTA GCT TAT CAG ACT G-3'; universal R: 5'-AGT GCA GGG TCC GAG G-3'.
U6: U6-RT: 5'-CGC TTC ACG AAT TTG CGT GTC-3'.
U6: F: 5'-TCG CTT CGG CAG CAC ATA TAC-3'; U6 R: 5'-GCG TGT CAT CCT TGC GCA G-3'.
Western blot analysis
Cells were lysed with radioimmunoprecipitation assay (RIPA) buffer (Beyotime Biotechnology, China), and total protein was quantified using a protein assay dye (Bio-Rad, CA, USA). The samples were mixed with 4× sample buffer and heated to 95 °C for 10 minutes, and then 20 µg of protein was separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After electrophoresis, proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane (Millipore, MA, USA). The PVDF membranes were first blocked for 1 hour in tris-buffered saline (TBS) containing 0.1% Tween 20 and 5% non-fat powdered milk, and then incubated with primary antibodies at 4 °C overnight. The PVDF membranes were washed three times with TBS containing 0.1% Tween 20, with 10 minutes between each wash, and incubated with secondary antibodies at room temperature for 1 hour. Protein bands were visualized with the chemiluminescent HRP substrate (Millipore, MA, USA). To ensure similar protein loading, the membrane was probed with a monoclonal antibody specific for GAPDH or actin.
Cell proliferation and viability assays
Cell proliferation was evaluated using the MTT assay. Different concentrations of drugs were used, ranging from 0.0316 to 3,160 µM of 5-FU for HCT116 and HCT116/FUR cells. Cells were seeded in 96-well micro-titer plates at an initial density of 8,000 cells/well. After incubation for 24 hours, cells were incubated with a serial dilution of 5-FU for 72 hours. Subsequently, 10 µL of MTT solution (1 mg/mL) was added to each well and incubated for 1 hour. The supernatant was then aspirated, and 150 µL of DMSO was added to each well. Absorbance was measured on a microculture plate reader at 490 nm. Data were analyzed using GraphPad Prism 8 (GraphPad Software Inc., CA, USA), and are represented as mean ± standard deviation (SD) from three independent experiments.
Colony formation assay
Cells were treated as mentioned in the results section. After 36 hours, cells were trypsinized with 0.05% trypsin and counted. A total of 800 cells were seeded in the 6-well plate and incubated at 37 °C with 5% CO2. After 2 weeks, cells were stained using crystal violet and the number of colonies was counted using public domain software (ImageJ).
Cytotoxicity assay
Cells were seeded in 12-well plates and treated as mentioned in the results. After 72 hours, plates were stained with crystal violet for 10 minutes and washed in running tap water. At room temperature, plates were then dried overnight. Crystal violet staining was then dissolved in 100 mM of freshly made sodium citrate solution for 1 hour. The colorimetric absorbance was measured at 590 nm. Data were analyzed using GraphPad Prism 8 and are represented as mean ± SD from three independent experiments.
Xenograft model
Cell line xenografts were established by subcutaneously injecting 2×106 HCT116 or HCT116/FUR cells into both flanks of BALB/c-nu/nu mice (female, 4±1 weeks of age, weighing 18±2 g; GemPharmatech Co, Ltd, China). Tumors were allowed to grow for 7 to 14 days. Tumor-bearing mice were then randomized into six groups (n=5/group) and subjected to the treatment regimens described below.
HCT116 group: (I) vehicle, treated with 4% DMSO in sterile saline (i.p.) and 4% DMSO in corn oil (i.g.); (II) 5-FU, treated with 40 mg/kg 5-FU (i.p.) and 4% DMSO in corn oil (i.g.).
HCT116-FUR group: (III) vehicle, treated with 4% DMSO in sterile saline (i.p.) and 4% DMSO in corn oil (i.g.); (IV) 5-FU, treated with 40 mg/kg 5-FU (i.p.) and 4% DMSO in corn oil (i.g.); (V) SP600125: treated with 15 mg/kg SP600125 (i.g.) and 4% DMSO in sterile saline (i.p.); (VI) Combo: treated with 40 mg/kg 5-FU (i.p.) and 15 mg/kg SP600125 (i.g).
SP600125 was dissolved in corn oil with 4% DMSO, while 5-FU was dissolved in DMSO and then diluted in sterile saline. Mice were treated for 5 days/week, for 2 weeks. Mice were accommodated in a specific pathogen-free (SPF)-grade animal room, and ethically sacrificed 2 weeks after treatment.
After 1 week of the treatment, 2 random mice from each group were ethically sacrificed, and the tumors were harvested for RT-qPCR analysis of abcg2. Tumors were measured by a caliper every other day, and the tumor volume was calculated as V = a×b2/2, where a represents the longer axis diameter and b is the shorter axis diameter.
Animal experiments were performed under a project license (No. 20191205001) granted by the Sichuan University Committee on Animal Care and Use, in compliance with the NIH Guide for the Care and Use of Laboratory Animals, and the Sichuan University Guide for the Care and Use of Laboratory Animals.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 8 and ImageJ. Data are presented as the mean ± SD for at least three individual experiments for each group. Statistical differences were determined using ANOVA and Student’s t-test for independent samples. A P value <0.05 was considered statistically significant.
Ethical statement
The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Results
ABCG2 conferred resistance to 5-FU in CRC cells
It has been reported that 5-FU is the direct substrate of ABCG2 in breast cancer cells (11). We found that ABCG2 expression was significantly upregulated upon 5-FU treatment in HCT116 cells (Figure 1A,B). To explore the role of ABCG2 in resistance to 5-FU in CRC cells, a 5-FU-resistant CRC HCT116/FUR cell line was established by continuous treatment with increasing doses of 5-FU for approximately 6 months, and the sensitivity of HCT116/FUR cells to 5-FU was evaluated using an MTT assay and colony formation assay. As shown in Figure 1C, compared to parental HCT116 cells, HCT116/FUR cells showed significantly enhanced resistance to 5-FU (the IC50 increased from 6.9 µM in parental HCT116 cells to 337 µM in HCT116/FUR cells). This result was further confirmed by the colony formation assay (Figure 1D) and crystal violet staining assay (Figure 1E). Subsequently, mRNA and protein expression of ABCG2 in both HCT116 and HCT116/FUR cells were assessed. We found that both mRNA and protein expression levels of ABCG2 in HCT116/FUR cells were higher than those in HCT116 cells (Figure 1F,G).
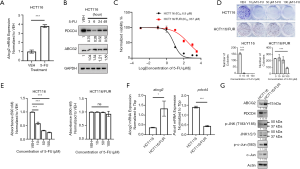
To investigate whether ABCG2 was involved in 5-FU resistance, HCT116 and HCT116/FUR cells were transfected with ABCG2 overexpression plasmids and siRNAs targeting ABCG2, respectively (Figure 2). As expected, overexpression of ABCG2 increased the 5-FU IC50 in HCT116 cells (Figure 2A), but silencing of ABCG2 by siRNAs decreased the 5-FU IC50 to a lesser extent in HCT116/FUR cells (Figure 2B). In addition, the results of the colony formation assay in HCT116 cells showed that silencing ABCG2 expression increased the sensitivity to 5-FU (Figure 2C). These results indicated that ABCG2 might confer partial 5-FU resistance in CRC cells.
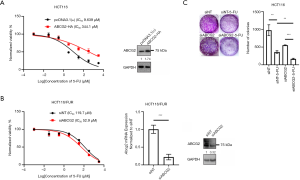
Inhibition of the JNK pathway attenuated ABCG2 and increased the sensitivity to 5-FU in HCT116 and HCT116/FUR cells in vitro
The JNK pathway has been shown to be involved in drug resistance, and can be exploited for overcoming P-glycoprotein-mediated MDR (15). We found that 5-FU treatment upregulated phosphorylation levels of c-Jun and JNK in parental HCT116 cells (Figure 3A). Compared to parental HCT116 cells, phosphorylation levels of c-Jun and JNK were also increased in HCT116/FUR cells (Figure 1G). Thus, we hypothesized that the JNK pathway mediated the increasing ABCG2 expression observed with 5-FU treatment in CRC cells. To verify this hypothesis, a JNK pathway inhibitor, SP600125, was employed to inhibit the kinase activity of JNK. As shown in Figure 3A, exposure of HCT116 and HCT116/FUR cells to SP600125 blocked the increased phosphorylation of c-Jun at serine 63, which confirmed the effectiveness of SP600125. Inhibition of the JNK pathway by SP600125 attenuated the 5-FU-induced upregulation of ABCG2 in both HCT116 and HCT116/FUR cells, especially in HCT116/FUR cells (Figure 3A,B). These results suggested that the JNK pathway mediated the increase in ABCG2 expression induced by 5-FU. To explore whether inhibition of the JNK pathway can enhance the sensitivity of HCT116 or HCT116/FUR cells to 5-FU, several assays were conducted. The colony formation assay showed that the number of colonies was reduced upon cotreatment with 5-FU and SP600125, compared to 5-FU treatment alone in HCT116/FUR cells (Figure 3C). This result was further confirmed by the MTT assay and crystal violet assay (Figure 3D,E). Overall, these results suggested that the JNK pathway conferred 5-FU resistance via upregulating ABCG2 in CRC cells, and inhibition of the JNK pathway increased the sensitivity of CRC cells to 5-FU.
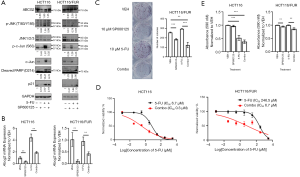
Repressing PDCD4 activated the JNK/ABCG2 pathway in HCT116/FUR cells
PDCD4 has been shown to suppress neoplastic transformation by reducing activator protein 1 (AP-1)-dependent transcription and inhibiting the expression of mitogen-activated protein kinase kinase kinase kinase 1 (MAP4K1) to block the activation of c-Jun (16). We found that PDCD4 expression was downregulated upon 5-FU treatment in HCT116 cells (Figure 1A), which was also verified in other CRC cells including RKO cells (Figure 4A). In addition, PDCD4 expression in HCT116/FUR cells was less than that in parental HCT116 cells (Figure 1F,G). In HCT116 and RKO cells, exogenous expression of PDCD4 blocked the phosphorylation of c-Jun and JNK and downregulated ABCG2 72 hours post-transfection (Figure 4B). Moreover, when PDCD4 was decreased by transient expression of shPDCD4 in HCT116 cells, ABCG2 mRNA was significantly upregulated (Figure 4C,D) through increasing the phosphorylation levels of JNK and c-Jun (Figure 4D). Furthermore, rescuing PDCD4 expression in HCT116/FUR cells restored sensitivity to 5-FU by repressing the JNK/ABCG2 pathway and inducing apoptosis (Figure 4E,F,G). These data indicated that decreased PDCD4 expression by 5-FU mediated the chemoresistance to 5-FU via activating the JNK/ABCG2 pathway in CRC cells.
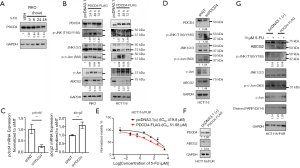
miR-21 mediated chemoresistance to 5-FU by regulating the PDCD4/JNK/ABCG2 pathway
PDCD4 has been shown to be a direct target of miR-21 (17). In the present study, miR-21 expression was found to be upregulated by 5-FU treatment in HCT116 and RKO cells (Figure 5A). In addition, miR-21 was significantly increased in HCT116/FUR cells (Figure 5B). To explore whether miR-21 mediates 5-FU-induced PDCD4 downregulation, we used RKO and RKO-KO cells as models and evaluated PDCD4 expression in both cell lines. As expected, both the mRNA and protein expression of PDCD4 in RKO-KO cells were much higher than those in the wild-type RKO cells (Figure 5C,D). Moreover, rescue of miR-21 expression by miR-21 nucleotide mimics decreased PDCD4 expression in RKO-KO cells (Figure 5E). These results suggested that miR-21 was responsible for the 5-FU-induced downregulation of PDCD4.
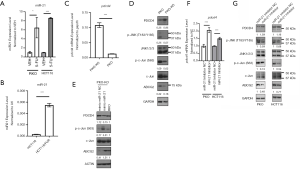
We then explored whether miR-21 mediates resistance to 5-FU through the PDCD4/JNK pathway. As shown in Figure 5D, phosphorylation levels of JNK and c-Jun and protein expression levels of ABCG2 were lower in RKO-KO cells compared to RKO cells. Exogenous miR-21 mimics significantly increased the phosphorylation level of c-Jun and ABCG2 expression (Figure 5E). In accordance with the results of RKO-KO cells, blocking miR-21 function by an miR-21 inhibitor upregulated the expression of PDCD4, and attenuated the phosphorylation levels of c-Jun and JNK, along with the expression of ABCG2 in both RKO and HCT116 cells (Figure 5F,G). Taken together, these results indicated that upregulation of miR-21 by 5-FU induced the partial resistance to 5-FU through regulating the PDCD4/JNK/ABCG2 pathway.
JNK inhibition sensitized HCT116/FUR cells to 5-FU in vivo
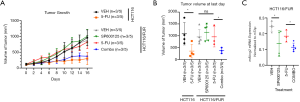
To investigate whether interfering with the JNK/ABCG2 pathway could recover sensitivity to 5-FU in vivo, HCT116 and HCT116/FUR-derived xenograft nude mouse models were established. The results showed that 5-FU significantly inhibited the tumor growth of parental HCT116 cells, but not HCT116/FUR cells (Figure 6A,B). Tumor volumes of the HCT116/FUR group with combination treatment of 5-FU and SP600125 treatment were significantly reduced compared to those of the HCT116/FUR group with 5-FU or SP600125 treatment alone (Figure 6A,B). In addition, the HCT116/FUR Combo (SP600125 and 5-FU) group showed decreased ABCG2 mRNA expression (Figure 6C). These results revealed that the administration of SP600125 recovered the sensitivity to 5-FU in 5-FU-resistant mice via inhibition of the JNK/ABCG2 pathway. The weights of each mice were monitored during the experiments (data not shown), and throughout the duration of the in vivo experiments. There were no notable adverse events among the mice across the experimental groups. These data corresponded to the results in vitro.
Discussion
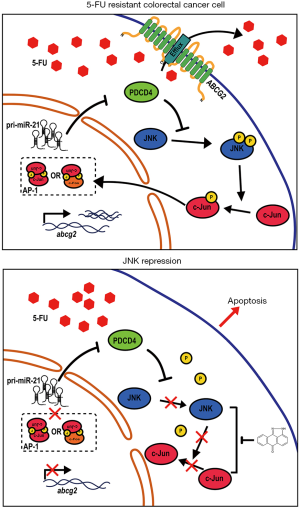
CRC is one of the most common malignant tumors worldwide. Even though morbidity and mortality of CRC have been slowly decreasing over the past several decades, the numbers of predicted new cases and deaths are still increasing in some areas (1). 5-FU-based chemotherapy is widely used for the treatment of CRC; however, the development of 5-FU resistance has been the main obstacle to successful CRC treatment. Although several underlying mechanisms of 5-FU resistance in cancer cells have been identified, its complexity necessitates further study to discover novel mechanisms. Some of the ABC transporter superfamily members have been recognized as efflux pumps for various chemotherapeutic compounds or their metabolites, consequently inducing drug resistance. It has been reported that ABCC5, ABCC11, and ABCG2 were directly involved in 5-FU resistance by the efflux transport of the active metabolites FdUMP or 5-FU (11,18,19). Nevertheless, the mechanisms underlying the involvement of ABCG2 in 5-FU resistance in CRC remain to be comprehensively understood. In the present study, ABCG2 expression was upregulated upon treatment with 5-FU in HCT116 cells. Of note, ABCG2 was found to degrade 72 hours after 5-FU treatment (data not shown). Peng et al. reported that the half-life of ABCG2 is approximately 54 hours (20). Furthermore, we found that a 72-hour duration of treatment caused cell death in most HCT116 cells. Therefore, we assumed that ABCG2 induced by 5-FU might degrade upon long-term treatment with 5-FU, and that most HCT116 cells with a low expression of ABCG2 would die. Long-term treatment with 5-FU might lead to epigenetic changes in cells resulting in highly expressed ABCG2 in 5-FU-resistant CRC cells (HCT116/FUR) without 5-FU treatment. In our study, 5-FU treatment in HCT116/FUR further induced ABCG2 expression. Furthermore, when exogenous ABCG2 was expressed in HCT116 parental cells, 5-FU sensitivity was inhibited. Loss of ABCG2 expression by siRNA targeting ABCG2 in HCT116 cells decreased clonogenic survival in HCT116 cells. However, silencing ABCG2 by transient transfection of siABCG2 in HCT116/FUR cells did not fully recover the sensitivity to 5-FU. Our findings further verified that acquired 5-FU resistance in HCT116 cells was partially associated with elevated expression of ABCG2. When HCT116 cells are treated with 5-FU, ABCG2 will accumulate in cells and pump the 5-FU out of cells to induce the acquired resistance (Figure 7).
JNK, also known as stress-activated MAP kinase (SAPK), is a member of the MAPK family, and has been shown to bind to the NH2-terminal activation domain of the transcription factor c-Jun (21). The JNK signaling pathway has been reported to be activated by a range of stimuli, including hormones, pathogens, cytokines, UV radiation, oxidative stress, and DNA damage (21). Upon activation, JNK binds to the activation domain of c-Jun and subsequently phosphorylates serine 63 and serine 73 residues, which inhibits the degradation of c-Jun, and leads to increasing transcriptional activity of the AP-1 complex containing c-Jun and c-FOS (22). Over 100 proteins have been recognized as JNK substrates, including the anti-apoptotic protein Bcl-2 and the pro-apoptotic protein Bax (23). The JNK signaling pathway has been reported to regulate embryonic morphogenesis, cell proliferation, tumor transformation, and apoptosis (24-26). Additionally, its role in both pro- and anti-apoptosis depends on cell type, intensity and dose of the stimulus, duration of treatment, and the activities of other cellular signaling pathways (27-30). The role of the JNK pathway in drug resistance has been demonstrated in various cancer types, including colon cancer and lung adenocarcinoma. Overall, the JNK signaling pathway has been shown to be overactivated in most drug resistance cases. Recently, JNK1 has been reported to regulate ABCG2-induced resistance to 10-hydroxycamptothecin (HCPT) in colon cancer cells (14). In addition, Scotto et al. reported that AP-1 can bind to the promoter of ABCG2 and activate transcription of ABCG2 (31). We found that phospho-JNK, c-Jun, and phospho-c-Jun (Ser63) were increased in HCT116 cells treated with 5-FU, and upregulated in HCT116/FUR cells. Blocking the JNK pathway using the inhibitor SP600125 recovered the sensitivity to 5-FU in vitro and in vivo, and abolished overexpression of ABCG2 in HCT116/FUR cells. Recently, Yao et al. reported that the therapeutic efficacy of 5-FU was enhanced by augmented activation of the JNK pathway via overexpressed LAST2, confirming the pro-apoptotic role of the JNK pathway in reversing 5-FU resistance in CRC cells (32). The different functions of the JNK pathway in 5-FU-resistant CRC cells might be due to different cell lines or different stages of resistance, which requires further analyses using clinical tumor samples and in vitro and in vivo investigations.
PDCD4 is a novel tumor suppressor and has been found to have lost or reduced expression in several cancers, including colon cancer. Overexpressed PDCD4 was shown to selectively increase sensitivity of cisplatin, paclitaxel, and carboplatin (33-35). PDCD4 has also been reported to be inhibited by miRNA-21 or miRNA-302a to induce 5-FU resistance in hepatocellular carcinoma cells and pancreatic cancer cells (36-38). Studies have demonstrated that PDCD4 can inhibit AP-1-dependent transcription in a concentration-dependent manner via inhibition of the transactivation of MAP4K1, c-Jun, and possibly c-FOS (39,40). In this study, we found that PDCD4, whose expression was repressed in HCT116/FUR cells, modulated the JNK/ABCG2 pathway and thereby decreased the sensitivity to 5-FU, which further verified the role of PDCD4 in acquired resistance to 5-FU in CRC cells.
Previous studies have shown that miR-21 plays an important role in 5-FU resistance in colon cancer cells, and miR-21 overexpression dramatically reduces the therapeutic efficacy of 5-FU both in vitro and in vivo (41,42). In addition, PDCD4 has been identified as one of the main direct targets of miR-21 in CRC (17,43). In this study, miR-21 expression significantly increased after 5-FU treatment. In HCT116/FUR cells, miR-21 was also significantly increased. In addition, re-expression of miR-21 downregulated PDCD4 and activated the phosphorylation of c-Jun in RKO-KO cells. Furthermore, neutralization of miR-21 function by introduction of an miR-21 inhibitor in RKO and HCT116 cells attenuated the phosphorylation of JNK and c-Jun, and ABCG2 expression. All these results suggested that miR-21 partially mediated 5-FU-induced downregulation of PDCD4 and activation of the JNK/ABCG2 pathway. However, the detailed mechanisms require further investigation.
Collectively, our study showed that in 5-FU-resistant CRC cells, repression of PDCD4 activated the JNK pathway, which resulted in increased ABCG2 expression. Overexpression of ABCG2 on the membrane might efflux 5-FU, and subsequently induce acquired resistance to 5-FU (Figure 7A). When the JNK pathway was blocked, 5-FU resistance was overcome by diminishing ABCG2 overexpression (Figure 7B). Inhibition of the JNK pathway also enhanced the anti-tumor activity of 5-FU both in vivo and in vitro. Together, our results demonstrate the critical role of the miR-21/PDCD4/JNK/ABCG2 axis in acquired resistance to 5-FU in CRC cells. In turn, our data contributes towards a better understanding of 5-FU resistance mechanisms, and provides insights into overcoming 5-FU resistance by targeting the miR-21/PDCD4/JNK/ABCG2 axis.
Acknowledgments
We thank Prof. Lin Zhang at University of Pittsburgh for his helpful comments on the manuscript and project. We would also like to thank Mr. Eliot D. Lee at University of California at San Francisco for his comments on the manuscript.
Funding: This study was supported by the National Natural Science Foundation of China (#81672942).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-4292
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-4292
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-4292). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Animal experiments were performed under a project license (No. 20191205001) granted by Sichuan University Committee on Animal Care and Use, in compliance with the NIH Guide for the Care and Use of Laboratory Animals, and Sichuan University Guide for the Care and Use of Laboratory Animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Heidelberger C, Chaudhuri NK, Danneberg P, et al. Fluorinated pyrimidines, a new class of tumour-inhibitory compounds. Nature 1957;179:663-6. [Crossref] [PubMed]
- Wohlhueter RM, McIvor RS, Plagemann PG. Facilitated transport of uracil and 5-fluorouracil, and permeation of orotic acid into cultured mammalian cells. J Cell Physiol 1980;104:309-19. [Crossref] [PubMed]
- Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer 2002;2:48-58. [Crossref] [PubMed]
- Kugimiya N, Nishimoto A, Hosoyama T, et al. The c-MYC-ABCB5 axis plays a pivotal role in 5-fluorouracil resistance in human colon cancer cells. J Cell Mol Med 2015;19:1569-81. [Crossref] [PubMed]
- Xie T, Geng J, Wang Y, et al. FOXM1 evokes 5-fluorouracil resistance in colorectal cancer depending on ABCC10. Oncotarget 2017;8:8574-89. [Crossref] [PubMed]
- Hu H, Wang M, Guan X, et al. Loss of ABCB4 attenuates the caspase-dependent apoptosis regulating resistance to 5-Fu in colorectal cancer. Biosci Rep 2018;38:BSR20171428. [Crossref] [PubMed]
- Bessho Y, Oguri T, Achiwa H, et al. Role of ABCG2 as a biomarker for predicting resistance to CPT-11/SN-38 in lung cancer. Cancer Sci 2006;97:192-8. [Crossref] [PubMed]
- Bleau AM, Huse JT, Holland EC. The ABCG2 resistance network of glioblastoma. Cell Cycle 2009;8:2936-44. [Crossref] [PubMed]
- Chikazawa N, Tanaka H, Tasaka T, et al. Inhibition of Wnt signaling pathway decreases chemotherapy-resistant side-population colon cancer cells. Anticancer Res 2010;30:2041-8. [PubMed]
- Yuan J, Lv H, Peng B, et al. Role of BCRP as a biomarker for predicting resistance to 5-fluorouracil in breast cancer. Cancer Chemother Pharmacol 2009;63:1103-10. [Crossref] [PubMed]
- Yu B, Gu D, Zhang X, et al. The role of GLI2-ABCG2 signaling axis for 5Fu resistance in gastric cancer. J Genet Genomics 2017;44:375-83. [Crossref] [PubMed]
- Meyer zu Schwabedissen HE, Grube M, Dreisbach A, et al. Epidermal growth factor-mediated activation of the map kinase cascade results in altered expression and function of ABCG2 (BCRP). Drug Metab Dispos 2006;34:524-33. [Crossref] [PubMed]
- Zhu MM, Tong JL, Xu Q, et al. Increased JNK1 signaling pathway is responsible for ABCG2-mediated multidrug resistance in human colon cancer. PLoS One 2012;7:e41763. [Crossref] [PubMed]
- Zhou J, Liu M, Aneja R, et al. Reversal of P-glycoprotein-mediated multidrug resistance in cancer cells by the c-Jun NH2-terminal kinase. Cancer Res 2006;66:445-52. [Crossref] [PubMed]
- Yang HS, Matthews CP, Clair T, et al. Tumorigenesis suppressor Pdcd4 down-regulates mitogen-activated protein kinase kinase kinase kinase 1 expression to suppress colon carcinoma cell invasion. Mol Cell Biol 2006;26:1297-306. [Crossref] [PubMed]
- Allgayer H. Pdcd4, a colon cancer prognostic that is regulated by a microRNA. Crit Rev Oncol Hematol 2010;73:185-91. [Crossref] [PubMed]
- Oguri T, Bessho Y, Achiwa H, et al. MRP8/ABCC11 directly confers resistance to 5-fluorouracil. Mol Cancer Ther 2007;6:122-7. [Crossref] [PubMed]
- Pratt S, Shepard RL, Kandasamy RA, et al. The multidrug resistance protein 5 (ABCC5) confers resistance to 5-fluorouracil and transports its monophosphorylated metabolites. Mol Cancer Ther 2005;4:855-63. [Crossref] [PubMed]
- Peng H, Qi J, Dong Z, et al. Dynamic vs static ABCG2 inhibitors to sensitize drug resistant cancer cells. PLoS One 2010;5:e15276. [Crossref] [PubMed]
- Weston CR, Davis RJ. The JNK signal transduction pathway. Curr Opin Cell Biol 2007;19:142-9. [Crossref] [PubMed]
- Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell 2000;103:239-52. [Crossref] [PubMed]
- Zeke A, Misheva M, Reményi A, et al. JNK Signaling: Regulation and Functions Based on Complex Protein-Protein Partnerships. Microbiol Mol Biol Rev 2016;80:793-835. [Crossref] [PubMed]
- Ding Q, Wang Q, Evers BM. Alterations of MAPK activities associated with intestinal cell differentiation. Biochem Biophys Res Commun 2001;284:282-8. [Crossref] [PubMed]
- Endo H, Hosono K, Fujisawa T, et al. Involvement of JNK pathway in the promotion of the early stage of colorectal carcinogenesis under high-fat dietary conditions. Gut 2009;58:1637-43. [Crossref] [PubMed]
- Tong C, Yin Z, Song Z, et al. c-Jun NH2-terminal kinase 1 plays a critical role in intestinal homeostasis and tumor suppression. Am J Pathol 2007;171:297-303. [Crossref] [PubMed]
- Hess P, Pihan G, Sawyers CL, et al. Survival signaling mediated by c-Jun NH(2)-terminal kinase in transformed B lymphoblasts. Nat Genet 2002;32:201-5. [Crossref] [PubMed]
- Papachristou DJ, Batistatou A, Sykiotis GP, et al. Activation of the JNK-AP-1 signal transduction pathway is associated with pathogenesis and progression of human osteosarcomas. Bone 2003;32:364-71. [Crossref] [PubMed]
- Potapova O, Basu S, Mercola D, et al. Protective role for c-Jun in the cellular response to DNA damage. J Biol Chem 2001;276:28546-53. [Crossref] [PubMed]
- Yang YM, Bost F, Charbono W, et al. C-Jun NH(2)-terminal kinase mediates proliferation and tumor growth of human prostate carcinoma. Clin Cancer Res 2003;9:391-401. [PubMed]
- Scotto KW. Transcriptional regulation of ABC drug transporters. Oncogene 2003;22:7496-511. [Crossref] [PubMed]
- Yao W, Zhu S, Li P, et al. Large tumor suppressor kinase 2 overexpression attenuates 5-FU-resistance in colorectal cancer via activating the JNK-MIEF1-mitochondrial division pathway. Cancer Cell Int 2019;19:97. [Crossref] [PubMed]
- Jansen AP, Camalier CE, Stark C, et al. Characterization of programmed cell death 4 in multiple human cancers reveals a novel enhancer of drug sensitivity. Mol Cancer Ther 2004;3:103-10. [PubMed]
- Shiota M, Izumi H, Tanimoto A, et al. Programmed cell death protein 4 down-regulates Y-box binding protein-1 expression via a direct interaction with Twist1 to suppress cancer cell growth. Cancer Res 2009;69:3148-56. [Crossref] [PubMed]
- Zhang X, Wang X, Song X, et al. Programmed cell death 4 enhances chemosensitivity of ovarian cancer cells by activating death receptor pathway in vitro and in vivo. Cancer Sci 2010;101:2163-70. [Crossref] [PubMed]
- Tomimaru Y, Eguchi H, Nagano H, et al. MicroRNA-21 induces resistance to the anti-tumour effect of interferon-α/5-fluorouracil in hepatocellular carcinoma cells. Br J Cancer 2010;103:1617-26. [Crossref] [PubMed]
- Wang W, Zhao L, Wei X, et al. MicroRNA-320a promotes 5-FU resistance in human pancreatic cancer cells. Sci Rep 2016;6:27641. [Crossref] [PubMed]
- Wei X, Wang W, Wang L, et al. MicroRNA-21 induces 5-fluorouracil resistance in human pancreatic cancer cells by regulating PTEN and PDCD4. Cancer Med 2016;5:693-702. [Crossref] [PubMed]
- Wang Q, Zhang Y, Yang HS. Pdcd4 knockdown up-regulates MAP4K1 expression and activation of AP-1 dependent transcription through c-Myc. Biochim Biophys Acta 2012;1823:1807-14. [Crossref] [PubMed]
- Yang HS, Knies JL, Stark C, et al. Pdcd4 suppresses tumor phenotype in JB6 cells by inhibiting AP-1 transactivation. Oncogene 2003;22:3712-20. [Crossref] [PubMed]
- Valeri N, Gasparini P, Braconi C, et al. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2). Proc Natl Acad Sci U S A 2010;107:21098-103. [Crossref] [PubMed]
- Deng J, Lei W, Fu JC, et al. Targeting miR-21 enhances the sensitivity of human colon cancer HT-29 cells to chemoradiotherapy in vitro. Biochem Biophys Res Commun 2014;443:789-95. [Crossref] [PubMed]
- Li S, Liang Z, Xu L, et al. MicroRNA-21: a ubiquitously expressed pro-survival factor in cancer and other diseases. Mol Cell Biochem 2012;360:147-58. [Crossref] [PubMed]
(English Language Editors: C. Betlazar-Maseh and J. Gray)


