
Predicting aggressive histopathological features in esophageal cancer with positron emission tomography using a deep convolutional neural network
Introduction
Esophageal cancer is one of the most common causes of cancer-related death worldwide. In 2012, there were an estimated 455,800 new esophageal cancer cases and 400,200 deaths due to esophageal cancer worldwide (1). Even in the early stages, treatment failure is common after radical surgery such as transthoracic esophagectomy with extended lymph node dissection, and the prognosis remains poor (2). Multidisciplinary treatment, including combinations of chemotherapy, radiotherapy, and surgical resection, has been introduced to reduce systemic micrometastasis and increase the complete resection rate in locally advanced esophageal cancer. However, a significant proportion of patients still experience disease recurrence after trimodal treatment (3). Measurements of esophageal cancer aggressiveness are primarily based on the anatomical extent of the disease, including tumor length, depth, and involvement of lymph node or distant organs, which are obtained from clinical examinations, such as computed tomography, esophagogastroscopy, and esophageal endoscopic ultrasound (4). Non-anatomic factors, such as the presence of lymphovascular invasion (LVI) and perineural invasion (PNI), have also been shown to have a large impact on patient survival; however, such information is not available before tumor specimens are collected by surgical resection (5,6). The only non-anatomical assessment of esophageal cancer that can be obtained prior to surgical resection is positron emission tomography (PET), a nuclear medicine imaging technique based on the measurement of gamma rays emitted by a positron-emitting radiotracer, such as 18F-fludeoxyglucose (FDG) (7). Generally, FDG uptake is represented as a standardized uptake value, which measures the highest image pixel in each tumor region. FDG uptake is able to assess metabolic activity and can localize the primary tumor as well as any metastases that may be present. Although PET is currently the only means of functional assessment prior to treatment, whether PET results correlate with prognostic histopathological features remains to be elucidated.
Deep neural networks, in particular convolutional neural networks (CNNs), have been increasingly applied to medical image analysis for image classification, image regression, object detection, and image segmentation. The CNN is image-based machine learning. The errors caused by inaccurate feature calculation and segmentation can be avoided and performance can be higher than that of ordinary feature-based classifiers (8). In landmark studies, Gulshan et al. (9), Esteva et al. (10), and Ehteshami Bejnordi et al. (11) demonstrated the potential of deep learnings CNNs to detect diabetic retinopathy, classify skin lesions, and diagnosis lymph node metastasis in breast cancer, respectively. Furthermore, CNNs can be trained to recognize biological features that are overlooked by human experts (12,13). In our previous study, we showed that a CNN can be trained with PET image datasets to predict esophageal cancer outcome with acceptable accuracy (14). In the present study, we trained a CNN using PET images to predict the presence of LVI/PNI. We aim to evaluate whether deep learning CNN can unlock hidden information in PET scans and connect functional images with histopathological features. We present the following article in accordance with the TRIPOD reporting checklist (available at http://dx.doi.org/10.21037/atm-20-1419).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Institutional Review Board of Taipei-Veterans General Hospital and granted a waiver of the informed consent process (IRB 2018-01-019-AC).
Data source and preprocessing
The PET scans of patients with the diagnosis of esophageal squamous cell carcinoma between September 2009 and August 2017 at Taipei Veterans General Hospital were collected. The PET scans of patients with stage I lung cancer diagnosed between January 2012 and November 2017 at Taipei Veterans General Hospital were also collected for use in first part of this study to pretrain the neural network. Clinicopathological information was gathered from a prospectively established database. The pixel resolution of each PET scan in XY was 128×128, with a pixel size of 5.47 mm × 5.47 mm and a slice thickness of 3.27 mm. Raw PET images in DICOM format were first converted to standardized uptake value images. To focus on critical information, we cropped images to remove any irrelevant areas. Each cropped PET scan covered the body area from the hypopharynx to the stomach and included all of the esophagus and the peri-esophageal regions. The field of view of each cropped scan was 32×32×128 pixels. Additionally, we increased the effective size of the training set using on-the-fly data augmentation. Six augmentation methods were employed in this study: random image translation, random scaling, random rotation, random left/right flipping, random swapping of anterior/posterior view, and random Gaussian blurring. Finally, each input image was zero-centered by subtracting the mean and the min/max normalized by dividing the image by its intensity range.
Model setup
Clinical factors were classified as binary values. Input images with tumors were classified as positive if they showed either LVI or PNI by post-surgical histopathological examination, while those without LVI or PNI were labeled as negative. We built a three-dimensional (3D)-CNN based on a residual network, ResNet (15). For our neural network, we applied full pre-activation, which reorganizes the order of convolutions and activation functions so that batch normalization and a rectified linear unit (ReLU) precede convolution layer. This results in better performance compared to original residual block or other configurations, such as ReLU before addition or ReLU-only pre-activation (16). After several convolutions, we used a global averaging method to flatten the extracted features as a vector. This vector was then connected to the dense layer using a softmax function, with the output being the probability of a given image being classified as positive for either LVI or PNI or negative for both LVI and PNI.
Hyper-parameters setting
The kernel weights of the network were initialized using a recipe published by He et al. (17). To train the model, we used Stochastic Gradient Descent (SGD) (18-20) with Nesterov momentum (21,22) (with an initial learning rate of 0.0001 and momentum of 0.9). We applied a Bayesian optimization method for searching several hyper-parameters combinations, including batch size, learning rate, with training and validation set. According to results of Bayesian optimization, the batch size was set to 12 and the learning rate was set to 9e-4. After hyper-parameters optimization, we trained our model with the same hyper-parameters combination and evaluated the testing set on a single nVIDIA TESLA P40 GPU. Since the model would over-fit easily if it was trained on an unbalanced dataset, we used a batch balancing method, which mixes over-sampling and under-sampling to circumvent class imbalance in the dataset (23). This method allowed us to train the model with balanced samples (6 positive and 6 negative samples per batch). To further prevent model over-fitting, reduce-learning-rate-on-plateau and early-stopping were included during the training process. The reduce-learning-rate-on-plateau schema generally dropped the learning rate by half when a validation performance did not improve for a few epochs, and early-stopping caused the model to stop training when a validation performance did not show improvement after 25 epochs.
Statistics analysis
To evaluate the performance in classifying esophageal cancer with specific aggressive histopathological features, we randomly split 80%, 10%, and 10% of our dataset into training set, validation set and testing set, respectively. We tuned the hyper-parameters with training set and validation set via the Bayesian optimization process. The model performance was evaluated by the testing set in the end.
Samples with predicted probability for each classification were split into either positive or negative using various thresholds ranging from 0 to 1 and the model performance was mainly evaluated by the area under receiver operating characteristic (ROC) curve (AUC). To assess statistical metrics, such as sensitivity, specificity, precision, recall, F1-score, and accuracy, we trained the model with same parameters 30 times after parameter searching. Hence, each sample in the testing set can be evaluated multiple times and derived the models’ statistical metrics.
Results
Patients
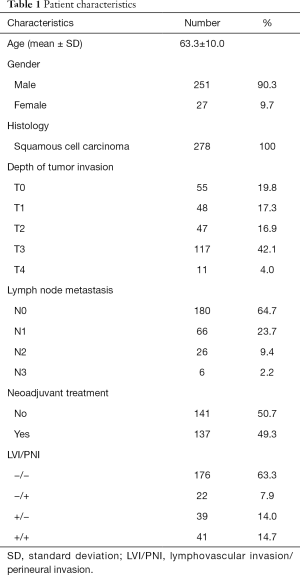
A total of 798 PET scans from 548 patients with diagnosis of esophageal squamous cell carcinoma and another 309 PET scans from patients with stage I lung cancer were included in the first part of this study. The demographic characteristics of total enrolled patients with esophageal squamous cell carcinoma was shown in our previous report (14). Among the esophageal cancer patients, 278 patients, who underwent esophagectomy for esophageal cancer, were included in the second part of this study for training a deep learning classifier to predict presence or absence of LVI/PNI by their PET scans (Figure 1).
Performance of classifying lung and esophageal cancer
We built a 3D-CNN as illustrated in Figure 2. In the first part of this study, which aimed to classify PET scans into esophageal cancer or lung cancer, the model showed an AUC of 0.5 and an accuracy of 0.717 under random guessing, in which the parameters of the neural network were set to random values. Our first part training converged after about 2,700 iterations and attained over 0.860 AUC, 0.867 F1-score, and 0.811 accuracy. The sensitivity was 0.850, and the specificity was 0.710. This result shows that our model has the ability to extract important features that can differentiate different cancers.
Performance of classifying esophageal cancer with specific histopathological features
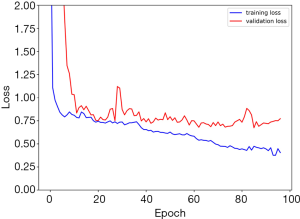
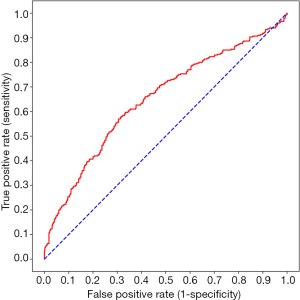
In the second part, the demographic characteristics of the 278 patients with esophageal squamous cell carcinoma are listed in Table 1. The median interval between PET scan and esophagectomy was 10 days (Q1: 5; Q3: 22 days). Our 3D-CNN model, more specifically, an 18-layer ResNet with the SGD optimizer, was trained for classifying PET scans into presence or absence of LVI/PNI. We randomly spilt dataset into training set, validation set and testing set with 80%, 10%, 10%, respectively. Models generally converged after about 1,600 iterations and attain 0.668 AUC (w/o pretrain) and 0.660 AUC (w/pretrain) on average. The learning curves of the networks showed that the training and validation loss generally decrease and converge along with training time (Figure 3). The other statistical results on the testing set of our 3D-CNN’s ability to classify patients by the presence or absence of LVI/PNI based on a threshold of 0.5 are listed in Table 2. In the ROC curve analysis, the highest combination of sensitivity and specificity occurred at a sensitivity of 0.574 and a specificity of 0.663, based on a predicted probability threshold of 0.45 (Figure 4).
Full table
Full table
Discussion
CNNs are a class of deep learning methods that perform especially well on image recognition tasks. A CNN is composed of multiple network layers, such as convolution layers, pooling layers, and fully connected layers; automatically extracts key features from a training data set; and adaptively learns spatial hierarchies of features through a backpropagation algorithm as well as by fine-tuning the hyperparameters of each neural network layer (24). The application of CNN in clinical medical imaging is rapidly increasing. For a topic with an abundance of well-annotated data, the performance of CNN can be outstanding. For example, classification of skin cancer (10), classification of dermoscopic melanoma recognition (25), detection of diabetic retinopathy with retinal fundus photographs (9), detection of lymph node metastases with whole slide images in breast cancer (11), and anatomical classification of esophagogastroduodenoscopy images (26) have been performed using CNN, with resulting AUCs ranging from 0.79 to 0.99. Intriguingly, it has been proposed that CNN may be able to astutely reveal subtle biological characteristics that are not visible to physicians. As examples, researchers have used CNN to predict survival in colorectal cancer with haematoxylin-eosin-stained tumor tissue (27) and to predict cardiovascular risk factors with retinal fundus photographs (13). The resulting discrimination abilities have been acceptable or excellent; for example, an AUC of 0.7 for predicting major cardiac events. Few studies have applied CNN with PET imaging in thoracic oncology. In one study, CNN was trained to classify mediastinal lymph nodes (positive or negative) of non-small cell lung cancers from FDG-PET images (28). In another study, Ypsilantis et al. (29) used data from 107 patients with esophageal cancer to demonstrate that CNN has the potential to extract PET imaging representations that are highly predictive of response to chemotherapy.
To our knowledge, this is the first report to apply CNN to predict specific histopathological features in esophageal cancer. The significance of LVI and PNI has been well-established in the literature (5,6), and patients with these features have been shown to have worse outcomes and a higher risk of recurrence after treatment. However, information about these two histological features is currently impossible to gather prior to major surgical resection. Whereas most clinical examinations for esophageal cancer evaluate tumor behavior based on the anatomical extent of disease, PET scans measure the metabolic activity of the tumor. Although relationships between functional imaging and immunohistochemical biomarkers have been reported (30), no study has investigated the association between PET scans and specific histological characteristics in esophageal cancer. In this study, deep learning was applied in an attempt to unlock “hidden” information in PET scans and connect functional images with histological features. One caveat of this deep learning approach is the necessity of a large dataset to train the vast number of parameters contained in a CNN. For target datasets that are considered too small to successfully train a CNN, it is a common practice to use a large image dataset of similar physical characteristics (imaging modality) and image content (for example, natural objects) but of different object category to pre-train a network before finally training the network with the target dataset. Previous studies showed that finetuning networks based on pretrained weights, such as ImageNet, generally get better or equal results (31). Because there is no large 3D dataset of medical images available for pre-training 3D CNNs, we conducted a two-stage workflow in which the weights of network were pretrained based on a differentiating normal and abnormal esophagus task before being tasked with classifying image data based on whether LVI/PNI were present in the abnormal esophagus. Our results showed that pre-training generally didn’t show significant difference compared to training without pre-trained weights. It may indicate either learned features do not transfer well to our main task or the pre-trained weights were still limited by a relative small dataset compared to the data size of ImageNet.
This study is novel because it explored the possible links between PET scan data and histological examination results. Our 3D-CNN used PET images of the entire esophagus as input, which eliminated the need for PET slice selection, tumor segmentation, and feature selection. To overcome the limitations of a small dataset and the lack of a 3D image dataset for transfer learning, we adopted a stepwise workflow. This study is also limited by the fact that the histological examinations were based on hematoxylin and eosin staining without special markers, e.g., CD34 and podoplanin for LVI or S100 to detect nerve fibers (32,33). The actual LVI/PNI percentage may have thus differed if more specific immunohistochemistry had been performed.
To conclude, our 3D-CNN can be trained with PET imaging datasets to predict LNV/PNI in esophageal cancer with acceptable accuracy. Although our current results cannot be readily applied to clinical decision making, we demonstrated the potential of deep learning to uncover hidden information in PET scans and connect functional imaging with histopathological findings. With a larger dataset, the CNN can be trained to achieve a better prediction performance.
Acknowledgments
This study has been presented in the poster session (PS02.070) of 16th ISDE World Congress, 2018.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-1419
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-1419
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-1419). JCYY is the CEO of aetherAI, Co., Ltd. WHY and CKY are the employees of aetherAI, Co., Ltd. PKH serves as a current Editorial Team member. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Institutional Review Board of Taipei-Veterans General Hospital and granted a waiver of the informed consent process (IRB 2018-01-019-AC).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Jang R, Darling G, Wong RK. Multimodality approaches for the curative treatment of esophageal cancer. J Natl Compr Canc Netw 2015;13:229-38. [Crossref] [PubMed]
- Hsu PK, Chen HS, Huang CS, et al. Patterns of recurrence after oesophagectomy and postoperative chemoradiotherapy versus surgery alone for oesophageal squamous cell carcinoma. Br J Surg 2017;104:90-7. [Crossref] [PubMed]
- Udagawa H, Ueno M. Comparison of two major staging systems of esophageal cancer-toward more practical common scale for tumor staging. Ann Transl Med 2018;6:76. [Crossref] [PubMed]
- Lagarde SM, Phillips AW, Navidi M, et al. The presence of lymphovascular and perineural infiltration after neoadjuvant therapy and oesophagectomy identifies patients at high risk for recurrence. Br J Cancer 2015;113:1427-33. [Crossref] [PubMed]
- Hsu PK, Chien LI, Wang LC, et al. Lymphovascular invasion and extracapsular invasion are risk factors for distant recurrence after preoperative chemoradiotherapy and esophagectomy in patients with esophageal squamous cell carcinoma. Eur J Cardiothorac Surg 2017;51:1188-94. [Crossref] [PubMed]
- Goel R, Subramaniam RM, Wachsmann JW. PET/Computed tomography scanning and precision medicine: Esophageal Cancer. PET Clin 2017;12:373-91. [Crossref] [PubMed]
- Suzuki K. Overview of deep learning in medical imaging. Radiol Phys Technol 2017;10:257-73. [Crossref] [PubMed]
- Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 2016;316:2402-10. [Crossref] [PubMed]
- Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115-8. [Crossref] [PubMed]
- Ehteshami Bejnordi B, Veta M, Johannes van Diest P, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 2017;318:2199-210. [Crossref] [PubMed]
- Maxmen A. Deep learning sharpens views of cells and genes. Nature 2018;553:9-10. [Crossref]
- Poplin R, Varadarajan AV, Blumer K, et al. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat Biomed Eng 2018;2:158-64. [Crossref] [PubMed]
- Yang CK, Yeh JC, Yu WH, et al. Deep convolutional neural network-based positron emission tomography analysis predicts esophageal cancer outcome. J Clin Med 2019;8:844. [Crossref] [PubMed]
- He K, Zhang X, Ren S, et al. Deep residual learning for image recognition (2015). Available online: https://arxiv.org/pdf/1512.03385.pdf
- He K, Zhang X, Ren S, et al. Identity mappings in deep residual networks (2016). Available online: https://arxiv.org/pdf/1603.05027.pdf
- He K, Zhang X, Ren S, et al. Delving deep into rectifiers: surpassing human-level performance on imagenet classification (2015). Available online: https://arxiv.org/pdf/1502.01852.pdf
- Robbins H, Monro S. A stochastic approximation method. Ann Math Statist 1951;22:400-7. [Crossref]
- Kiefer J, Wolfowitz J. Stochastic estimation of the maximum of a regression function. Ann Math Statist 1952;23:462-6. [Crossref]
- Bottou L, Curtis FE, Nocedal J. Optimization methods for large-scale machine learning (2018). Available online: https://arxiv.org/pdf/1606.04838.pdf
- Nesterov YE. A method for unconstrained convex minimization problem with the rate of convergence o(1/k2). Dokl Akad Nauk SSSR 1983;269:543-7.
- Sutskever I, Martens J, Dahl G, et al. On the importance of initialization and momentum in deep learning. Proceedings of the 30th International Conference on Machine Learning, PMLR 2013;28:1139-47.
- Hernandez J, Carrasco-Ochoa JA, Martínez-Trinidad JF. An empirical study of oversampling and undersampling for instance selection methods on imbalance datasets. In: Ruiz-Shulcloper J, Sanniti di Baja G, eds. Progress in Pattern Recognition, Image Analysis, Computer Vision, and Applications. New York, NY: Springer; 2013.
- Yamashita R, Nishio M, Do RKG, et al. Convolutional neural networks: an overview and application in radiology. Insights Imaging 2018;9:611-29. [Crossref] [PubMed]
- Haenssle HA, Fink C, Schneiderbauer R, et al. Man against machine: diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann Oncol 2018;29:1836-42. [Crossref] [PubMed]
- Takiyama H, Ozawa T, Ishihara S, et al. Automatic anatomical classification of esophagogastroduodenoscopy images using deep convolutional neural networks. Sci Rep 2018;8:7497. [Crossref] [PubMed]
- Bychkov D, Linder N, Turkki R, et al. Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci Rep 2018;8:3395. [Crossref] [PubMed]
- Wang H, Zhou Z, Li Y, et al. Comparison of machine learning methods for classifying mediastinal lymph node metastasis of non-small cell lung cancer from 18 F-FDG PET/CT images. EJNMMI Res 2017;7:11. [Crossref] [PubMed]
- Ypsilantis PP, Siddique M, Sohn HM, et al. Predicting response to neoadjuvant chemotherapy with pet imaging using convolutional neural networks. PLoS One 2015;10:e0137036. [Crossref] [PubMed]
- Incoronato M, Grimaldi AM, Cavaliere C, et al. Relationship between functional imaging and immunohistochemical markers and prediction of breast cancer subtype: a PET/MRI study. Eur J Nucl Med Mol Imaging 2018;45:1680-93. [Crossref] [PubMed]
- Kornblith S, Shlens J, Le QV, Do Better ImageNet Models Transfer Better? Computer Vision and Pattern Recognition (2018). Available online: https://arxiv.org/abs/1805.08974
- Wang S, Chen X, Fan J, et al. Prognostic Significance of Lymphovascular Invasion for Thoracic Esophageal Squamous Cell Carcinoma. Ann Surg Oncol 2016;23:4101-9. [Crossref] [PubMed]
- Xu G, Feng F, Liu Z, et al. Prognosis and Progression of ESCC Patients with Perineural Invasion. Sci Rep 2017;7:43828. [Crossref] [PubMed]




