
The GABRG2 F343L allele causes spontaneous seizures in a novel transgenic zebrafish model that can be treated with suberanilohydroxamic acid (SAHA)
Introduction
Early onset epileptic encephalopathies (EOEEs) are a group of devastating epilepsy syndromes characterized by severe, treatment-resistant early onset seizures in the setting of global neurodevelopmental delay (1). EOEEs have a broad phenotypic spectrum and are highly heterogeneous at the molecular level (2). With the development of next-generation sequencing techniques, the role of genetic predisposition with de novo mutations is now increasingly recognized in EOEEs (3). Moreover, variants in ion channel genes are among the most prevalent causes of EOEEs (4). Recently, with whole exome sequencing in a patient with an EOEE, we identified a de novo mutation, c.T1027C, in the GABAA receptor gamma 2 subunit gene (GABRG2) that produced mutant γ2(F343L) subunits (5). Our in vitro studies in transfected HEK293T cells demonstrated that when coexpressed with α1 and β2 subunits, the mutant γ2(F343L) subunit impaired GABAA receptor biogenesis and channel function, suggesting the pathological basis for this mutation causing EOEE. To further validate the pathogenesis of the mutant γ2(F343L) subunit, we sought a model system that could facilitate studies of this mutation in vivo.
Considerable attention has focused on engineering knock in mice bearing epilepsy-associated GABRG2 mutations. However, mouse models are not high throughput models, are expensive to maintain, and have epilepsy phenotypes that are strongly influenced by background genetics (6). Danio rerio (zebrafish) has recently emerged as a powerful vertebrate model for studying epilepsy-related mutations with significant advantages for easy genetic manipulation, cost-efficient breeding and small size for high throughput drug screening (7). Within three to five days post fertilization (dpf), zebrafish develop into freely swimming larvae with a complex nervous system, and they can be evaluated for behavioral changes. Treatment of zebrafish larvae with the GABAA receptor antagonist convulsant drug pentylenetetrazol (PTZ) evoked stereotypical, dose-dependent seizure-like phenotypes, which correlated with epileptiform discharges in the forebrain and optic tectum of mutant zebrafish as evidenced by electrophysiological field potential recordings (8). Subsequent studies over the past decade have successfully reproduced these locomotor convulsive behaviors in genetically modified zebrafish to understand the function of epilepsy-related genes (9). Moreover, the usefulness of them for in vivo discovery of novel “precision medicine-focused” therapeutics has become increasingly apparent within the last few years (9,10).
Here, we established novel transgenic zebrafish models overexpressing either human wild-type γ2L or mutant γ2L(F343L) subunits to investigate the functional consequences of the patient’s mutant γ2(F343L) subunit in vivo. The expression level of the neuronal activity marker gene c-fos was significantly higher in the brain of mutant Tg(hGABRG2F343L) larvae than of wild-type Tg(hGABRG2wt) larvae. Video observation and locomotion tracking analysis at 5 dpf demonstrated the occurrence of seizure-like behaviors in mutant Tg(hGABRG2F343L), but not wild-type Tg(hGABRG2) larvae. Using RNA sequencing, we investigated transcriptomic changes due to the presence of mutant γ2(F343L) subunits and have found 524 genes that are differentially expressed compared to control, with 342 genes up-regulated and 182 genes down-regulated. In consistent with the impaired biogenesis and trafficking of mutant GABAA receptors, we identified a specific up-regulation of 33 genes involved in endoplasmic reticulum and protein processing pathway in Tg(hGABRG2F343L) brains. Protein network analysis indicated histone acetylation as a key modifier of the disrupted protein processing pathway, which might contribute to epileptogenesis.
Suberanilohydroxamic acid (SAHA, also known as Vorinostat) is an orally available histone deacetylases (HDACs) inhibitor that was approved by the US Food and Drug Administration for management of cutaneous T cell lymphoma (11). In vitro SAHA treatment has previously been shown to effectively enhance proteostasis, and thereby, partially correct the impaired function of GABAA receptors produced by epilepsy mutations associated with endoplasmic reticulum (ER) -associated protein degradation (12-14). Since SAHA crosses the blood brain barrier, we investigated its anticonvulsant efficacy and that of several established anticonvulsant drugs (valproic acid, levetiracetam, clonazepam, and carbamazepine) in vivo using the Tg(hGABRG2F343L) and Tg(hGABRG2wt) zebrafish models. Locomotion assays demonstrated that SAHA, but not the established anticonvulsant drugs, was effective at reducing seizure-like behaviors in a dose-dependent manner, supporting the potential utility of SAHA for the treatment of genetic epilepsies that result from GABAA receptor misfolding and by extension from misfolding of other missense mutant proteins.
We present the following article in accordance with the ARRIVE reporting checklist (available at: http://dx.doi.org/10.21037/atm-20-3745).
Methods
The animal experiments in this study were approved by the Ethics Committee of Nantong University (Approval ID: 20180202-001), in compliance with Nantong University guidelines for the care and use of animals.
Bioinformatics
Human GABRG2 (NM_000816.3) information was obtained from the GenBank database. GABAA receptor subunit sequences were aligned by the T-Coffee program (http://tcoffee.crg.cat/apps/tcoffee/do:regular) (15).
Zebrafish lines and breeding
The zebrafish (AB strain) embryos and adults were maintained in the Zebrafish Center of Nantong University under conditions in accordance with our previous protocols (16). All protocols and procedures were conducted in accordance with the institutional animal care guidelines of Nantong University. Adult wild-type zebrafish were maintained at 28.5 °C in a 14-hr light/10-hr dark cycle. PTU (1-phenyl-2-thio-urea), a commonly used tyrosinase inhibitor to block pigmentation and aid visualization of zebrafish development, was added to the embryos after 24 hours post fertilization (hpf). Zebrafish embryos and larvae were raised in 10-cm plastic petri dishes in an incubator maintained at 28.5 °C. Embryos were raised in E3 medium, containing (in mM) 5.0 NaCl, 0.17 KCl, 0.33 MgSO4, 0.33 CaCl2 and 0.2 PTU.
Transgene constructs and generation of transgenic zebrafish
Transgenic zebrafish were created using the Tol2kit transgenesis system and Gateway vectors. p5E-Huc was purchased from Addgene (Plasmid #72640) and originally from the Josh Bonkowsky Lab (17). The hGABRG2wt and hGABRG2F343L DNA sequences were synthesized by Union-Biotech (Shanghai) and subcloned into pME-MCS plasmid. The p3E-polyA plasmid was obtained from Tol2Kit (18). Three entry clones and the pDestTol2CG2 destination vector were used to generate the expression construct by LR recombination reaction as described in the Multisite Gateway Manual (Life Technologies). The green heart was used for the transgene screening. The synthetic Tol2 transposase mRNA, and the expression construct were co-injected into zebrafish embryos to efficiently generate germline integrations. Injected embryos were screened through examining fluorescence expression using stereo fluorescence microscopy. F0 founders were raised until sexually mature and individually mated with wild-type fish, and the offspring were examined for fluorescence expression in the heart. F1 embryos were obtained from one of the positive founders and raised.
RNA preparation, reverse transcription and quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted from zebrafish embryos by TRIzol reagent (Invitrogen, USA), and 2 µg total RNA was reversely transcribed using an Omniscript RT kit (Qiagen, USA) according to the manufacturer’s instructions. qRT-PCR was performed using the corresponding primers (Table 1) in a 20 µL reaction volume with 10 µL SYBR premix (Takara, Japan) and β-actin was used as the internal control. Data were normalized to the internal reference gene β-actin and quantified relative to the c-fos expression in Tg(hGABRG2wt) larvae at 24 hpf or GABRG2 expression in Tg(hGABRG2wt) larvae at 72 hpf. Values represent averages from four independent biological samples (one sample = ten pooled larvae) for each of the five developmental stages (24, 48, 72, 96 and 120 hpf).

Full table
Whole mount in situ hybridization
Whole mount in situ hybridization was performed as Thisse's protocol with modifications (19). A 767 bp cDNA fragment of c-fos was amplified from genomic DNA of wild-type AB embryos using specific primers (Table 1). Digoxigenin (DIG)-labeled sense and antisense probes were synthesized using linearized pGEM-T easy vector subcloned with the c-fos fragment by in vitro transcription with DIG-RNA labeling Kit (Roche, Switzerland). Zebrafish larvae were collected and fixed with 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) for one night. The fixed samples were then dehydrated through a series of increasing concentrations of methanol and stored at −20 °C in 100% methanol eventually. Hybridization was performed as previously described (16,20).
Behavioural assays in zebrafish larvae
Zebrafish larvae were raised in a 24-well-culture plate with one larva in each well filled with 1 mL E3 medium. On 5 dpf, larvae were transferred to the DanioVision Video-Track system (Noldus, German) (8). After adapting for 30 min, the larvae were videotaped with every 1 min forming a movement distance and trajectory by EthoVision XT locomotion tracking software version (Noldus, German). Seizure score, total distance traveled (mm), acceleration maximum (in mm per second square), and movement/no movement time (seconds) were noted and analyzed. Ten-min recording epochs were obtained for Tg(hGABRG2F343L) larvae (n=67) and age matched Tg(hGABRG2wt) siblings (n=68) bathed in normal embryo media at 5 dpf. Light stimulation was performed by switching between darkness (5 s) and light (5 s) for 100 cycles. Blinded analysis of the percentage of animals that reached each score based on the criteria previously described was performed. 28-36 larvae of each group were used for the locomotor activities assays.
Transcriptomic assay, differential expression assay and pathway analysis
Two independent batches of 5dpf Tg(hGABRG2wt) and Tg(hGABRG2F343L) larvae were dissected to extract the whole brains. Total RNA of each sample was extracted using TRIzol Reagent (Invitrogen, CA, USA), quantified and qualified by Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA), NanoDrop (Thermo Fisher Scientific, MA, USA) and 1% agrose gel. 1 µg total RNA with RIN value above 6.5 was used for following library preparation. Next generation sequencing library preparations were constructed according to the manufacturer’s protocol. Libraries with different indices were multiplexed and loaded on an Illumina HiSeq instrument according to manufacturer’s instructions (Illumina, CA, USA). Sequencing was carried out using a 2×150 bp paired-end (PE) configuration; image analysis and base calling were conducted by the HiSeq Control Software (HCS) + OLB + GAPipeline-1.6 (Illumina) on the HiSeq instrument. The sequences were processed and analyzed by GENEWIZ (Suzhou, China). Differential expression analysis used the DESeq2 Bioconductor package, a model based on the negative binomial distribution. The estimates of dispersion and logarithmic fold changes incorporate data-driven prior distributions, adjusted P value (Padj) of genes were set <0.05 to detect differential expressed ones. GOSeq (v1.34.1) was used to identify Gene Ontology (GO) terms that annotate a list of enriched genes with a significant Padj less than 0.05. Scripts in house was used to enrich significant differential expression genes in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways.
Drug administration
The following drugs were dissolved in dimethyl sulfoxide (DMSO, Sigma-Aldrich) to stock solutions, and stored at −20 °C: SAHA, valproic acid (VPA), clonazepam, levetiracetam, carbamazepine (all from Sigma-Aldrich). For drug administration, drugs in DMSO were diluted in E3 medium to a final concentration at 0.1, 0.5 and 2.5 µM for SAHA, 50 µM for VPA, 50 µM for clonazepam, 30 mM for levetiracetam and 100 µM for carbamazepine, respectively. Transgenic zebrafish larvae were randomly divided into different groups and 20 larvae of each group were used. Zebrafish larvae were incubated in E3 medium containing the drugs or DMSO for 30 min before transfer to the DanioVision Video-Track system, and the locomotion was recorded for 30 min.
Statistical analysis
Statistical analysis was performed by GraphPad Prism 6 software (GraphPad Software Inc., San Diego, CA, USA), and data were presented as means ± SEM. Comparisons were analyzed by Student’s two-tailed unpaired t-test or one-way analysis of variance (ANOVA) and subsequent Tukey’s multiple comparisons test between each group. Statistical significance was set as P<0.05.
Results
Tg(hGABRG2F343L) zebrafish larvae were generated
The zebrafish genome encodes orthologues for all but two of the nineteen human GABAA receptor subunit isoforms (21). A single orthologue of the human GABRG2 is localized on chromosome 21 of the zebrafish genome (NM_001256250.1). The invariant polar F343 residue is located in the deeper portion of the transmembrane α-helices M3 of the γ2 subunit, towards the +/- subunit interfaces. Of note, alignment analysis demonstrated that the zebrafish γ2 subunit is 97% identical to the human γ2 subunit (Figure 1A), supporting the relevance of investigating the functional consequences of the F343L mutation in γ2 subunits in zebrafish.
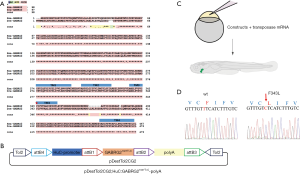
We introduced human wild-type and mutant GABRG2 transgenes into the zebrafish genome and generated transgenic zebrafish employing the Tol2kit transgenesis system and Gateway vectors. The expression of hGABRG2 [Tg(hGABRG2wt) or Tg(hGABRG2F343L)] was driven by the neuronal specific HuC promoter (Figure 1B). The expression constructs were microinjected with transposase mRNA into one-cell stage zebrafish embryos. The cmlc2: EGFP, which labels the embryonic heart with EGFP, was used for screening embryos with human GABRG2 expression (Figure 1C). We confirmed the genomic sequence by Sanger sequencing (Figure 1D). Mutant Tg(hGABRG2F343L) larvae did not exhibit any gross dysmorphologies compared to control larvae. Because genetic epilepsy is inherited most often in an autosomal dominant fashion, even a single mutant allele in the presence of the two wild-type alleles is likely to at least partially recapitulate the disease phenotype in transgenic fish.
Seizure-related c-fos gene transcripts were upregulated in Tg(hGABRG2F343L) larvae
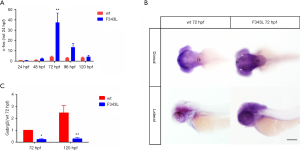
Vigorous and rapid induction of a program of immediate-early gene transcription in central nervous system is a hallmark of seizure onset in vertebrates. Seizure-induced expression of the synaptic-activity-dependent neuroprotective transcription factor c-fos has been observed in the brain of PTZ-treated larval zebrafish (8,22). We thus investigated the temporal expression of c-fos in transgenic zebrafish using qRT-PCR at several stages of early development, including 24, 48, 72, 96 and 120 hpf. At 72 hpf, RNA samples extracted from pooled Tg(hGABRG2F343L) larvae exhibited significantly higher levels of c-fos expression than with control larvae (Figure 2A). At 48, 96 and 120 hpf, c-fos expression in Tg(hGABRG2F343L) larvae was also higher compared to control siblings, although this was not statistically significant. Spatial patterns for the c-fos expression assessed using whole-mount in situ hybridization were consistent with PCR findings. C-fos was expressed at a relatively low level throughout the brain of Tg(hGABRG2wt) zebrafish larvae at 72 hpf. In age-matched Tg(hGABRG2F343L) transgenic larvae, a diffuse and prominent up-regulation of c-fos was observed in major central nervous system structures including telencephalon, optic tectum, midbrain, and hindbrain (Figure 2B). qRT-PCR also revealed a decrease in mRNA expression for GABRG2 in mutant larvae at 72 and 120 hpf compared to the wild-type transgenic line (Figure 2C).
Tg(hGABRG2F343L) zebrafish larvae exhibited spontaneous seizure-like activity
A seizure scoring system has been devised to provide an objective way of evaluating behavioral changes and categorize seizures in zebrafish larvae (8,23). Normal movement can be characterized as little (stage 0) or a general increase in swimming activity (stage 1), whereas seizure behaviors characterized as rapid “whirlpool-like” circling around the edge of the well (stage 2) and a series of high-speed whole-body convulsions culminating in a brief loss of posture (stage 3). Ten-min recording epochs were obtained for Tg(hGABRG2F343L) larvae (n=67) and age matched Tg(hGABRG2wt) siblings (n=68) bathed in normal embryo media at 5 dpf. Blinded analysis of the percentage of animals that reached each score based on the criteria above was shown (Figure 3A). As expected, there was no evidence of any seizure-like behaviors in Tg(hGABRG2wt) larvae, which only displayed behaviors characterized as stage 0 and 1. Of note, seizure-like behaviors consistent with stage 2 (22%, n=15) and stage 3 (6%, n=4) were exclusively observed in Tg(hGABRG2F343L) larvae. Examples of normal behaviors in Tg(hGABRG2wt) siblings and representative stage 2 and 3 seizure-like behaviors in the Tg(hGABRG2F343L) larvae were shown in Video S1.
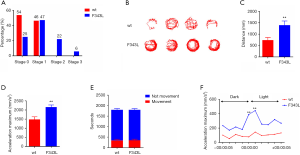
Increased locomotor activities are indicative of neuronal hyperexcitability and thus can serve as surrogate readouts for seizures (24,25). We next monitored the locomotor patterns of 5 dpf Tg(hGABRG2F343L) larvae and their siblings using an automated recording chamber. Representative swimming tracks of Tg(hGABRG2F343L) larvae showed hyperactivity (Figure 3B). We also evaluated locomotor behaviour by quantifying several behavioural parameters of the two transgenic lines. Tg(hGABRG2F343L) larvae traveled a significantly greater distance than Tg(hGABRG2wt) larvae (Figure 3C). Moreover, the acceleration maximum achieved by Tg(hGABRG2F343L) zebrafish was significantly higher compared to the control group (Figure 3D). Movement and no movement durations were relatively similar in both transgenic lines (Figure 3E). Upon light stimulation, we noticed that the acceleration maximum in Tg(hGABRG2F343L) larvae increased at the exact time the light was switched on, indicating a hypoactive response to light exposure (Figure 3F). Increased total locomotor activity and acceleration maxima were indicative of seizure activity in Tg(hGABRG2F343L) zebrafish.
GABRG2(F343L) mutation altered brain transcriptome of Tg(hGABRG2F343L) fish
To gain insights into the molecular defects and pathogenesis, we dissected whole brains from Tg(hGABRG2F343L) and Tg(hGABRG2wt) embryos, and extracted total RNA for deep sequencing. There was excellent concordance between transcriptional profiles of biological replicates. We used DESeq2 to analyze the differentially expressed genes amongst the 31,624 genes analyzed between the two groups. The transcriptome analysis identified 524 genes that were differentially expressed, including 342 transcripts whose abundance was increased ≥2-fold and 182 transcripts whose abundance was reduced by ≥2-fold in Tg(hGABRG2F343L) embryos when compared to the wild-type control (in total online: https://cdn.amegroups.cn/static/public/10.21037atm-20-3745-1.xlsx). A volcano plot of 524 genes with ≥2-fold changes in transcript abundance in transgenic fish was shown in Figure 4A, demonstrating broad changes in the transcriptome associated with the expression of GABRG2(F343L) mutation.
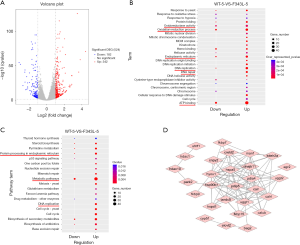
To investigate the function of the differentially expressed genes, GO enrichment analysis and KEGG pathway annotation were performed. GO analysis showed that these differentially expressed genes (DEG) were enriched for ATP binding, oxidoreductase activity, oxidation-reduction process in biological processes, for endoplasmic reticulum in cellular components, for DNA replication, DNA repair and protein folding in molecular functions (Figure 4B, in total online: https://cdn.amegroups.cn/static/public/10.21037atm-20-3745-2.xlsx). A more informative analysis of gene clustering was obtained using DAVID tool and identified the following clusters: metabolism (85 genes), cell-cell interaction (38 genes), DNA replication and repair (31 genes), mitochondria and endoplasmic reticulum (26 genes), protein processing (18 genes), neuronal function (8 genes) and development (4 genes). Notably, the DEG subnetwork is enriched in metabolic pathways, protein processing in endoplasmic reticulum and DNA replication (Figure 4C).
Brain transcriptome analysis identified HDACs as potential therapeutic targets in Tg(hGABRG2F343L) fish
Our in vitro studies in transfected HEK293T cells demonstrated that when coexpressed with α1 and β2 subunits, mutant γ2(F343L) subunits were retained in the endoplasmic reticulum, thereby disrupting the protein homeostasis network. Consistently, we identified a specific up-regulation of 33 genes involved in endoplasmic reticulum and protein processing pathway in Tg(hGABRG2F343L) brains (in total online: https://cdn.amegroups.cn/static/public/10.21037atm-20-3745-1.xlsx). Among these DEGs, hsp90b1, fkbp9, pdia6, txndc5, fkbp7, dnajb11, pdia4, bag2, hspd1, calr3b and calr3a are involved in protein folding. ube2t, uhrf1, park2, hspa5, uggt1 and uggt2 are related to protein ubiquitination and degradation. hdac1, hdac10 and hdac12 are responsible for protein acetylation. To detect the interrelationship between these DEGs, we constructed a protein-protein interaction (PPI) network (Figure 4D). Notably, hdac1 and hdac10 appear to work upstream in this network. HDAC10 has been linked to cellular stress response, protein degradation, autophagy and DNA repair (26). Indeed, several recent discoveries demonstrated that histone deacetylase inhibitors (HDACis) could modulate the deacetylation of chaperones, modify protein degradation pathways, and were effective in treating genetic diseases with misfolded proteins (27). Herein, we hypothesized that HDACis might be potential avenue for the treatment of epilepsy associated with GABAA receptor misfolding in Tg(hGABRG2F343L) larvae.
HDACs inhibitor SAHA reduced seizures in Tg(hGABRG2F343L) larvae
A decrease in locomotion in epileptic zebrafish is thought to be an indirect indicator of a reduction of seizure activity (28). As the patient carrying GABRG2(F343L) mutation remained intractable despite of combination therapy with available antiepileptic drugs (AEDs), we treated 5 dpf Tg(hGABRG2F343L) zebrafish larvae with a broad HDACs inhibitor, SAHA and then monitored its effect on spontaneous seizures using automated locomotion tracking software. We tested three drug concentrations based on a previous report (29) and our pilot experiments: 0.1 µM, 0.5 µM and 2.5 µM. Larvae locomotion was recorded for 30 min after a drug exposure of 30 min. Prolonged exposures were toxic at higher concentrations. SAHA treatment led to a significant decrease of total distance traveled in the Tg(hGABRG2F343L) zebrafish with a clear concentration-response relationship (Figure 5A,B).
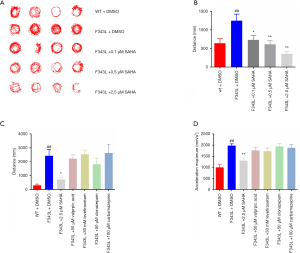
Additionally, we compared pharmaco-sensitivity of SAHA and four commercially available AEDs, valproic acid, levetiracetam, clonazepam, and carbamazepine (Figure 5C,D). Neither total distance nor acceleration maxima values were significantly changed for a 30-minute exposure to any of these traditional AEDs. In contrast, locomotion tracking data on SAHA-exposed Tg(hGABRG2F343L) zebrafish larvae confirmed a prominent reduction in these behavioural parameters, suggesting potential anti-seizure efficacy of SAHA.
Discussion
Establishment of a novel transgenic zebrafish model expressing the human GABRG2(F343L) mutation associated with EOEE
We recently reported finding a de novo GABRG2 mutation, F343L, in a human associated with an EOEE. In vitro, when co-transfected with α1 and β2 subunits in HEK293T cells, we found that the mutant subunit caused a reduction of receptor trafficking to the cell surface and retention of subunits in the ER, resulting in reduction of peak GABA-evoked current. To study this mutation in a vertebrate organism in vivo, we developed a novel transgenic zebrafish model bearing this mutation. Expression constructs were microinjected with transposase mRNA into one-cell stage zebrafish embryos using a transgene to produce overexpression of wild-type GABRG2 or mutant GABRG2(F343L). After injection, the wild-type larvae would have 3 wild-type copies of GABRG2 and the mutant larvae would have 2 wild-type copies and 1 mutant copy. The decreased expression of mutant GABRG2 in transgenic zebrafish line might be subject to nonsense-mediated messenger RNA decay. Mutant Tg(hGABRG2F343L) zebrafish exhibited seizure-like phenotypes in the larval stage, supporting the epileptic potential of the GABRG2(F343L) mutation in the affected child. Moreover, the hyperactivity induced upon light exposure indicated the phenotypes in mutant Tg(hGABRG2F343L) zebrafish could be triggered by light stimulus.
Our Tg(hGABRG2F343L) zebrafish exhibited a pharmacological profile like the drug-resistant epilepsies in humans. Seizure-like phenotypes in Tg(hGABRG2F343L) larvae were not responsive to acute (30 min dosing regimen) treatment with several commercially available AEDs. However, prolonged exposure of these AEDs for 24 h did appear to produce antiepileptic effects (data not shown). This mild effect of traditional AEDs was consistent with previous studies in a Scn1a mutant zebrafish model of Dravet syndrome (10,30).
Differential enrichment for genes involved in protein processing in Tg(hGABRG2F343L) zebrafish
To the best of our knowledge, this is the first study of transcriptome-wide expression changes in a zebrafish model of monogenic human epilepsy. Recently, a zebrafish homozygous GABRG2 null model was reported to display a mild phenotype of light/dark-induced reflex seizures and almost no change at the transcriptomic level (31). In contrast, our transcriptomic assay revealed significantly altered gene profiles in Tg(hGABRG2F343L) zebrafish brains.
Of note, many key genes involved in endoplasmic reticulum and protein processing pathway, most of which are molecular chaperones or co-chaperones, were markedly upregulated in Tg(hGABRG2F343L) brain (Figure 4D). For example, calr3a and calr3b encoding calreticulin, which assists the folding of the misfolded glycoprotein; pdia4 and pdia6 encoding PDI family, which inhibits the aggregation of misfolded proteins (32); uggt1 and uggt2 encoding UGGT, which re-glucosylates misfolded proteins (33); hspa5, encoding BiP protein, which is important in activation of unfolded protein response (UPR); dnajb11 and bag2 encoding co-chaperones of hsp40 or hsp70, which is required for the proper folding and assembly of proteins in the ER (34,35); ube2t, uhrf1 and park2; encoding E3 ubiquitin ligases, all involved in protein degradation in proteasomes or lysosomes (36-38). Notably, the expression of Hsp90β1 (also known as endoplasmin or grp94), an important chaperone protein in ER, was significantly increased. Up-regulation of Hsp90β has previously been identified in reactive astrocytes of the epileptic hippocampus in patients with temporal lobe epilepsy, and inhibition of hsp90β has been suggested as a potential therapeutic strategy for epilepsy (39). Altogether, these results strongly suggested that GABRG2(F343L) mutation disrupted proteostasis network in Tg(hGABRG2F343L) zebrafish brains.
Thus, the GABRG2(F343L) mutation may induce EOEE through both gain-of-function (related to the metabolism of mutant γ2 subunit protein) and loss-of-function mechanisms. Indeed, previous studies of genetic mouse models have shown that the pathogenesis of GABRG2 mutations is due to a combination of functional haploinsufficiency, dominant negative suppression as well as disturbance of cellular homeostasis related to metabolism of the mutant protein (40,41).
HDACs inhibition as a potential therapeutic paradigm for genetic epilepsies associated with missense GABAA receptor mutations
Although genetically modified mice harboring known GABRG2 mutations and exhibiting epilepsy have been developed, AEDs are rarely tested in these animals. In the present study, transcriptomic assay of our Tg(hGABRG2F343L) zebrafish model of epilepsy indicated histone acetylation as a key modifier of the disrupted protein processing pathway, which might contribute to epileptogenesis. Of interest from a therapeutic point of view, we subsequently translated this finding by demonstrating the efficacy of SAHA, a potent HDACs inhibitor, in alleviating seizure-like activities in Tg(hGABRG2F343L) zebrafish.
Histone modifications play a key role in the development of epilepsy, and HDACis have been successful in treating experimental epilepsy models (42,43). More recently, selective HDAC1 and HDAC3 inhibition have been shown to demonstrate potent anti-seizure effects in genetic epilepsy models of Kcna1-morpholino zebrafish larvae and Kcna1-null mice (29). As a broad HDACs inhibitor, SAHA has been shown to protect against seizure-induced brain damage through histone acetylation regulation (44). Intriguingly, SAHA was reported to enhance epilepsy-associated mutant GABAA receptor proteostasis in vitro (12-14). It has been demonstrated that SAHA increased the BiP protein level and the interaction between the mutant α1 subunit and calnexin, and thus partially corrected the transcription, folding and trafficking of mutant GABAA receptors (12). Here we speculated that the therapeutic effect of SAHA in our Tg(hGABRG2F343L) zebrafish was associated with modulation of cellular proteostasis due in part to activation of chaperone-mediated refolding, as reported in other human misfolding disorders (e.g., cystic fibrosis) (45).
Future research on elucidating the mechanism by which HDAC inhibition regulates the activity of chaperone proteins and the protein processing pathway is required. Additionally, we may proceed to test selective HDACis using our Tg(hGABRG2F343L) zebrafish model in an attempt to reduce side-effects.
Acknowledgments
Funding: This work was supported by National Natural Science Foundation of China (81801281, 81771404), Natural Science Foundation of Jiangsu Province (Grant No. BK20201440), Science and Technology Program of Nantong (MS22019002) and Jiangsu Provincial Key Medical Center and Jiangsu Students’ innovation and entrepreneurship training program (201913993005Y).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. available at: http://dx.doi.org/10.21037/atm-20-3745
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-3745
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure forms (available at: http://dx.doi.org/10.21037/atm-20-3745). The authors have no potential conflicts of interest to disclose.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The animal experiments in this study were approved by the Ethics Committee of Nantong University (Approval ID: 20180202-001), in compliance with Nantong University guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Scheffer IE, Berkovic S, Capovilla G, et al. ILAE classification of the epilepsies: Position paper of the ILAE Commission for Classification and Terminology. Epilepsia 2017;58:512-21. [Crossref] [PubMed]
- Shbarou R, Mikati MA. The Expanding Clinical Spectrum of Genetic Pediatric Epileptic Encephalopathies. Semin Pediatr Neurol 2016;23:134-42. [Crossref] [PubMed]
- Ellis CA, Petrovski S, Berkovic SF. Epilepsy genetics: clinical impacts and biological insights. Lancet Neurol 2020;19:93-100. [Crossref] [PubMed]
- Oyrer J, Maljevic S, Scheffer IE, et al. Ion Channels in Genetic Epilepsy: From Genes and Mechanisms to Disease-Targeted Therapies. Pharmacol Rev 2018;70:142-73. [Crossref] [PubMed]
- Shen D, Hernandez CC, Shen W, et al. De novo GABRG2 mutations associated with epileptic encephalopathies. Brain 2017;140:49-67. [Crossref] [PubMed]
- Maljevic S, Reid CA, Petrou S. Models for discovery of targeted therapy in genetic epileptic encephalopathies. J Neurochem 2017;143:30-48. [Crossref] [PubMed]
- Sakai C, Ijaz S, Hoffman EJ. Zebrafish Models of Neurodevelopmental Disorders: Past, Present, and Future. Front Mol Neurosci 2018;11:294. [Crossref] [PubMed]
- Baraban SC, Taylor MR, Castro PA, et al. Pentylenetetrazole induced changes in zebrafish behavior, neural activity and c-fos expression. Neuroscience 2005;131:759-68. [Crossref] [PubMed]
- Griffin A, Krasniak C, Baraban SC. Advancing epilepsy treatment through personalized genetic zebrafish models. Prog Brain Res 2016;226:195-207. [Crossref] [PubMed]
- Baraban SC, Dinday MT, Hortopan GA. Drug screening in Scn1a zebrafish mutant identifies clemizole as a potential Dravet syndrome treatment. Nat Commun 2013;4:2410. [Crossref] [PubMed]
- Richon VM. Targeting histone deacetylases: development of vorinostat for the treatment of cancer. Epigenomics 2010;2:457-65. [Crossref] [PubMed]
- Di XJ, Han DY, Wang YJ, et al. SAHA enhances Proteostasis of epilepsy-associated alpha1(A322D)beta2gamma2 GABA(A) receptors. Chem Biol 2013;20:1456-68. [Crossref] [PubMed]
- Chen X, Durisic N, Lynch JW, et al. Inhibitory synapse deficits caused by familial alpha1 GABAA receptor mutations in epilepsy. Neurobiol Dis 2017;108:213-24. [Crossref] [PubMed]
- Durisic N, Keramidas A, Dixon CL, et al. SAHA (Vorinostat) Corrects Inhibitory Synaptic Deficits Caused by Missense Epilepsy Mutations to the GABAA Receptor gamma2 Subunit. Front Mol Neurosci 2018;11:89. [Crossref] [PubMed]
- Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol 2000;302:205-17. [Crossref] [PubMed]
- Qin Y, Wang S, Duan X, et al. Expression analysis of the aquaporins during zebrafish embryonic development. Gene Expr Patterns 2019;32:38-43. [Crossref] [PubMed]
- Fujimoto E, Gaynes B, Brimley CJ, et al. Gal80 intersectional regulation of cell-type specific expression in vertebrates. Dev Dyn 2011;240:2324-34. [Crossref] [PubMed]
- Kwan KM, Fujimoto E, Grabher C, et al. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn 2007;236:3088-99. [Crossref] [PubMed]
- Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc 2008;3:59-69. [Crossref] [PubMed]
- Gong J, Chai L, Xu G, et al. The expression of natriuretic peptide receptors in developing zebrafish embryos. Gene Expr Patterns 2018;29:65-71. [Crossref] [PubMed]
- Cocco A, Ronnberg AM, Jin Z, et al. Characterization of the gamma-aminobutyric acid signaling system in the zebrafish (Danio rerio Hamilton) central nervous system by reverse transcription-quantitative polymerase chain reaction. Neuroscience 2017;343:300-21. [Crossref] [PubMed]
- Baxendale S, Holdsworth CJ, Meza Santoscoy PL, et al. Identification of compounds with anti-convulsant properties in a zebrafish model of epileptic seizures. Dis Model Mech 2012;5:773-84. [Crossref] [PubMed]
- Hortopan GA, Dinday MT, Baraban SC. Zebrafish as a model for studying genetic aspects of epilepsy. Dis Model Mech 2010;3:144-8. [Crossref] [PubMed]
- Pena IA, Roussel Y, Daniel K, et al. Pyridoxine-Dependent Epilepsy in Zebrafish Caused by Aldh7a1 Deficiency. Genetics 2017;207:1501-18. [Crossref] [PubMed]
- Zheng YM, Chen B, Jiang JD, et al. Syntaxin 1B Mediates Berberine's Roles in Epilepsy-Like Behavior in a Pentylenetetrazole-Induced Seizure Zebrafish Model. Front Mol Neurosci 2018;11:378. [Crossref] [PubMed]
- Ridinger J, Koeneke E, Kolbinger FR, et al. Dual role of HDAC10 in lysosomal exocytosis and DNA repair promotes neuroblastoma chemoresistance. Sci Rep 2018;8:10039. [Crossref] [PubMed]
- Yang C, Rahimpour S, Lu J, et al. Histone deacetylase inhibitors increase glucocerebrosidase activity in Gaucher disease by modulation of molecular chaperones. Proc Natl Acad Sci U S A 2013;110:966-71. [Crossref] [PubMed]
- Dinday MT, Baraban SC. Large-Scale Phenotype-Based Antiepileptic Drug Screening in a Zebrafish Model of Dravet Syndrome. eNeuro 2015;2:ENEURO.0068-15.2015.
- Ibhazehiebo K, Gavrilovici C, de la Hoz CL, et al. A novel metabolism-based phenotypic drug discovery platform in zebrafish uncovers HDACs 1 and 3 as a potential combined anti-seizure drug target. Brain 2018;141:744-61. [Crossref] [PubMed]
- Samarut E, Swaminathan A, Riche R, et al. gamma-Aminobutyric acid receptor alpha 1 subunit loss of function causes genetic generalized epilepsy by impairing inhibitory network neurodevelopment. Epilepsia 2018;59:2061-74. [Crossref] [PubMed]
- Liao M, Kundap U, Rosch RE, et al. Targeted knockout of GABA receptor gamma 2 subunit provokes transient light-induced reflex seizures in zebrafish larvae. Dis Model Mech 2019;12:dmm040782. [Crossref] [PubMed]
- Gorasia DG, Dudek NL, Safavi-Hemami H, et al. A prominent role of PDIA6 in processing of misfolded proinsulin. Biochim Biophys Acta 2016;1864:715-23. [Crossref] [PubMed]
- Roversi P, Marti L, Caputo AT, et al. Interdomain conformational flexibility underpins the activity of UGGT, the eukaryotic glycoprotein secretion checkpoint. Proc Natl Acad Sci U S A 2017;114:8544-9. [Crossref] [PubMed]
- Chen KC, Qu S, Chowdhury S, et al. The endoplasmic reticulum HSP40 co-chaperone ERdj3/DNAJB11 assembles and functions as a tetramer. EMBO J 2017;36:2296-309. [Crossref] [PubMed]
- Takayama S, Xie Z, Reed JC. An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperone regulators. J Biol Chem 1999;274:781-6. [Crossref] [PubMed]
- Cornwell MJ, Thomson GJ, Coates J, et al. Small-Molecule Inhibition of UBE2T/FANCL-Mediated Ubiquitylation in the Fanconi Anemia Pathway. ACS Chem Biol 2019;14:2148-54. [Crossref] [PubMed]
- Noda S, Sato S, Fukuda T, et al. Loss of Parkin contributes to mitochondrial turnover and dopaminergic neuronal loss in aged mice. Neurobiol Dis 2020;136:104717. [Crossref] [PubMed]
- Vaughan RM, Rothbart SB, Dickson BM. The finger loop of the SRA domain in the E3 ligase UHRF1 is a regulator of ubiquitin targeting and is required for the maintenance of DNA methylation. J Biol Chem 2019;294:15724-32. [Crossref] [PubMed]
- Sha L, Wang X, Li J, et al. Pharmacologic inhibition of Hsp90 to prevent GLT-1 degradation as an effective therapy for epilepsy. J Exp Med 2017;214:547-63. [Crossref] [PubMed]
- Warner TA, Shen W, Huang X, et al. Differential molecular and behavioural alterations in mouse models of GABRG2 haploinsufficiency versus dominant negative mutations associated with human epilepsy. Hum Mol Genet 2016;25:3192-207. [Crossref] [PubMed]
- Kang JQ, Macdonald RL. Molecular Pathogenic Basis for GABRG2 Mutations Associated With a Spectrum of Epilepsy Syndromes, From Generalized Absence Epilepsy to Dravet Syndrome. JAMA Neurol 2016;73:1009-16. [Crossref] [PubMed]
- Citraro R, Leo A, Santoro M, et al. Role of Histone Deacetylases (HDACs) in Epilepsy and Epileptogenesis. Curr Pharm Des 2017;23:5546-62. [Crossref] [PubMed]
- Reddy SD, Clossen BL, Reddy DS. Epigenetic Histone Deacetylation Inhibition Prevents the Development and Persistence of Temporal Lobe Epilepsy. J Pharmacol Exp Ther 2018;364:97-109. [Crossref] [PubMed]
- Hu QP, Mao DA. Histone deacetylase inhibitor SAHA attenuates post-seizure hippocampal microglia TLR4/MYD88 signaling and inhibits TLR4 gene expression via histone acetylation. BMC Neurosci 2016;17:22. [Crossref] [PubMed]
- Hutt DM, Herman D, Rodrigues AP, et al. Reduced histone deacetylase 7 activity restores function to misfolded CFTR in cystic fibrosis. Nat Chem Biol 2010;6:25-33. [Crossref] [PubMed]


