
Exploring plasma metabolomic changes in sepsis: a clinical matching study based on gas chromatography–mass spectrometry
Introduction
Sepsis is the second most common cause of mortality (>25%), and has a stable and persisting incidence in the intensive care unit (ICU) (1,2). The third international consensus conference for sepsis and septic shock (Sepsis-3) recently revised the definition of sepsis to include the key phrases: “life-threatening organ dysfunction” and “host response” (3). Studies have shown that infection involves extensive and complex pathogen–host interactions (4). These investigations revealed that sepsis develops as a result of amplification and subsequent dysregulation of the initial host response to an infection. Pathogens and their structural components thereby induce extensive changes in each system of the body including mitochondria-related metabolic system dysfunction (5). During sepsis-induced organ failure, inflammatory responses and oxidative stress induce changes in mitochondria, resulting in mitochondrial dysfunction and cell death. Thus, the “imbalance involving the internal environment” that causes energy metabolism disorders due to organ dysfunction is strongly associated with the host response (3).
Various biomarkers, such as procalcitonin and C-reactive protein, have been shown to be useful in the diagnosis and monitoring of sepsis (6). However, these biomarkers may also be elevated in other inflammatory environments such as trauma, burns, and in postoperative settings. In addition, sepsis also occurs as an end-stage characteristic of other conditions, including acute and chronic peritonitis, intravenous catheter infections, urinary tract stones, and diabetes, which also contributes to the low specificity of sepsis-related biomarkers (7). Therefore, identifying a novel sepsis biomarker may be difficult as some can reflect the nature of, and even respond to, dynamic changes during sepsis.
Metabolomics refers to all chemical reactions occurring within cells. It is considered a powerful platform for the global identification and quantification of low-molecular-weight metabolites in biological samples (8). Human metabolism directly responds to the microenvironment, and metabolite markers may represent multiple inputs (transcriptional, translational, and environmental) (6). Thus, metabolomics is particularly useful for the identification of sepsis-related biomarkers (as defined by Sepsis-3) by reflecting the internal environment. The present study conducted global metabolic profiling of the serum samples of patients with sepsis using gas chromatography–mass spectrometry (GC–MS). The patients were then further classified based on sepsis-related organ dysfunction to analyze metabolic differences in relation to organ dysfunction.
We present the following article in accordance with the Strengthening the Reporting of Observational studies in Epidemiology (STROBE) reporting checklist (available at http://dx.doi.org/10.21037/atm-20-3562).
Methods
Study design
The study was conducted in accordance with the World Medical Association’s Declaration of Helsinki (as revised in 2013), and was approved by the Ethics Committees of the First Affiliated Hospital of Chongqing Medical University (No. 2016-34). Informed consent was obtained from all patients. The patients included in this investigation were admitted to the ICU from October 2015 to February 2016, and the healthy control volunteers were from the Healthy Examination Center of the same medical institution. The healthy control volunteers had not been hospitalized, and physical and laboratory examinations were normal. The patients or their family members and the healthy control volunteers were fully informed of the study details and agreed to participate.
The inclusion criterion was a patient diagnosed with sepsis according to the Sepsis-3 definition 9 within 1 hour of hospitalization. The exclusion criteria were the following: (I) Patient’s with incomplete or inaccurate data. Since this study is a paired statistical analysis, complete data of each experimental object collected were required. If the data of the experimental object were incomplete, the collected samples could not be used in the range; (II) Patients diagnosed with chronic metabolic diseases.
According to Sepsis-3, the presence of organ dysfunction is also an important condition for the diagnosis of sepsis, and was a key diagnostic criterion in our study. For patients diagnosed with sepsis, plasma samples were collected within 1 hour of hospitalization, during the period before antibiotic treatment. The sepsis patients and healthy controls were matched in terms of age, body mass index (BMI), and gender. (Figure 1A) For each patient in this study, we used the Sequential Organ Failure Assessment (SOFA) score, quick SOFA (qSOFA) 9, and the Acute Physiology and Chronic Health Evaluation (APACHE) II and III scores 101112 to assess sepsis severity (Table 1).

Full table

Sample preparation and labeling for GC–MS
The serum samples were thawed on ice, and 100 µL of each sample were then diluted to a ratio of 1:1 with 100 µL of 0.2% formic acid solution, centrifuged at 14,000 rpm for 15 min at 4 °C, and placed on ice. Approximately 60 µL of each supernatant from the above two tubes were transferred into a new tube and immediately placed on ice. The samples were mixed and then placed in a −20 °C freezer. Prior to GC–MS analysis, the prepared samples were thawed on ice and centrifuged at a low speed to remove the foam and precipitate (Figure 1B).
GC–MS analysis
The samples were gasified on a vacuum centrifugal concentrator and analyzed by methyl chloride methyl ester (MCF). The program parameters were as follows: GCMS, ZB-1701 gas-phase capillary column (30 m × 320 µm × 0.25 µm, Phenomenex), inlet for Agilent splitter/splitless inlet; inlet temperature: 250 °C; FID detector temperature: 250 °C; nitrogen flow rate: 40 mL/min; hydrogen flow rate: 40 mL/min; airflow rate: 450 mL/min; split ratio: 20:1; sample injection volume: 1 µL. The program temperatures were as follows: an initial temperature of 80 °C, which was then increased at a rate of 25 °C/min to 200 °C, and then further increased at a rate of 3 °C/min to 215 °C, and finally increased at a rate of 2 °C/min to 230 °C for analysis (Figure 1B).
Subgroup data extraction (Figure 1A)
The acute kidney injury (AKI) (n=16) and non-AKI (n=15) subgroups were separated from the sepsis group based on criterion from the Kidney Disease Improving Global Outcomes (KDIGO) consensus conference, which defined markers of kidney damage or glomerular filtration rate (GFR) as <60 mL/min per 1.73 m2 for ≤3 months. The acute respiratory distress syndrome (ARDS) (n=22) and non-ARDS (n=9) subgroups were separated from the sepsis cohort based on the oxygenation index according to the Berlin ARDS definition: partial pressure of oxygen (PaO2)/fraction of inspired oxygen (FiO2) ≤300 mmHg. As there is no definitively agreed upon definition or criteria for sepsis-induced myocardial dysfunction (SIMD), diagnosis can be difficult. However, some studies have shown that troponin (cTn) and brain natriuretic peptide (BNP) have diagnostic and prognostic value in SIMD. We therefore separated SIMD (n=17) and non-SIMD (n=14) subgroups from the sepsis cohort based on the levels of myoglobin (MB), creatine kinase-myocardial band (CK-MB) isoenzyme, n-terminal pro-BNP (NT-proBNP), and troponin-I (cTnI) (9). Acute hepatic ischemia (AHI) associated with sepsis was defined by a serum bilirubin level greater than 2 mg/dL, with elevation of the serum glutamic oxaloacetic transaminase and lactic dehydrogenase levels about twice that of normal values 1,718. Therefore, the AHI (n=19) and non-AHI (n=12) subgroups were separated from the sepsis cohort based on serum bilirubin and amino-transferase levels (10). All plasma samples in each subgroup were matched to healthy volunteers in terms of age, BMI, and gender.
Data normalization
Automated Mass Spectral Deconvolution and Identification System (AMDIS) software was employed to deconvolute GC–MS chromatograms and identify metabolites using MCF mass spectra library 19. The identifications were based on the both MS spectrum of the derivatized metabolite and its respective chromatographic retention time. The relative abundance of identified metabolites was determined by ChemStation (Agilent) by using the GC base-peak value of a selected reference ion. These values were normalized using the biomass content in each sample and internal standard abundance.
Statistical analysis
Student’s t-test was used to determine whether the relative abundance of each identified metabolite was significantly different between sepsis samples and controls. The partial least squares discriminant analysis (PLS-DA) and receiver operating characteristic (ROC) curves were drawn using Microsoft Excel (Microsoft Corporation, USA) and inserted into Multibase (Digital Dynamics, Japan). Our Pathway Activity Profiling (PAPi) algorithm 20 was used to predict and compare the relative activity of different metabolic pathways in sepsis. This program is linked to the Kyoto Encyclopedia of Genes and Genomics (KEGG) online database (http://www.kegg.com) and uses the number of metabolites identified from each pathway and their relative abundances to predict which metabolic pathway is likely to be active in sepsis. The entire data mining, data normalization, and pathway activity predictions were automated by our in-house R software package. Graphical representations of the results were produced by ggplot2 R packages. In addition, correlation analysis of metabolites in various subgroups and clinical indicators was performed. Statistical analysis was performed using GraphPad Prism 6.0.
Results
Comparison of sepsis and control groups
General information on sepsis and control groups
Assessment of basic information indicated that there were no statistically significant differences in age, BMI, and gender between the sepsis and control groups (Table 1). Leave-one-out cross-validation (LOOCV) was used to verify the model (Figure 2A). The correlation coefficients (R2) and (Q2) of the established model were 0.74 and 0.75, respectively. PLS-DA indicated that there were significant differences between sepsis patients and healthy controls (Figure 2B).
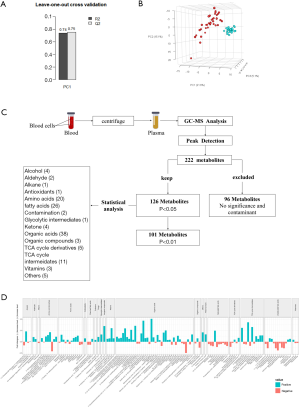
Analysis of metabolites between the sepsis patients and controls
The present study identified 222 metabolites, 124 of which showed statistical differences between the sepsis patients and healthy controls (Figure 2C,D). Of these, 26 were fatty acids, including 3 branched fatty acids, 10 saturated fatty acids, and 13 unsaturated fatty acids, which were found in sepsis plasma samples but not in the controls (Figure 2D). The 3 branched fatty acids included 3-methyl-2-oxopentanoic acid; 4-methyl-2-oxopentanoic acid; and tetradecanoic acid, 12-methyl, methyl ester. The (B) 10 saturated fatty acids included 10,13-dimethyltetradecanoic acid; arachidic acid; docosapentaenoic acid; hexanoic acid; margaric acid; nonadecanoic acid; palmitic acid; pentadecanoic acid; propanedioic acid, methyl, ethyl ester; and stearic acid. The 13 unsaturated fatty acids included 11,14-eicosadienoic acid; 11,14,17-eicosatrienoic acid; 2-methyloctadecanoic acid; 3-hydroxyoctanoic acid; adrenic acid; arachidonic acid; bishomo-gamma-linolenic acid; conjugated linoleic acid; docosahexaenoic acid (DHA); eicosapentaenoic acid (EPA); linoleic acid; myristoleic acid; and palmitoleic acid (Figure 2D). In addition, the levels of 20 of these 26 fatty acids (76.9%) were lower in the sepsis samples, with the 6 exceptions being tetradecanoic acid [12-methyl-, methyl ester, (S)-], hexanoic acid, 2-methyloctadecanoic acid, palmitoleic acid, myristoleic acid, and 3-hydroxyoctanoic acid (6/26; 23.1%). There were 12 amino acids in the differential metabolites, 7 of which (58.3%) were higher in the sepsis samples compared to the control, and included leucine, glutamic acid, cysteine, methionine, phenylalanine, putrescine, and aspartic acid. The levels of the five remaining amino acids [(including serine, tryptophan, glutamine, d-Proline, (N-methoxycarbonyl-, octyl ester) and asparagine] were lower in the sepsis samples compared to the control. However, the levels of six of the eight amino acid derivatives (75%) were higher in sepsis patients than in healthy volunteers. Also, the levels of 27 of the 38 (71.1%) differential organic acids (including lactic acid, adipic acid, and 3-hydroxypropionic acid) were higher in sepsis samples compared to the controls. Furthermore, the levels of pyruvic acid (one of the glycolytic intermediates) and nicotinamide adenine dinucleotide phosphate (NADP)-NADPH (one of the co-factors) were higher in the sepsis samples than in controls. We also found that five derivatives of the tricarboxylic acid (TCA) cycle showed differential levels compared to the control. Three of these (citraconic acid, citramalic acid, and DL-gamma-methyl-ketoglutaramate isomer 1) showed lower levels, whereas the levels of the remaining two (DL-gamma-methyl-ketoglutaramate isomer and dimethyl fumarate) were higher than the control. In addition, among the 11 TCA cycle intermediates in the sepsis samples, 7 differential metabolites were higher than those of the control (Figure 2C).
Pathway analysis of sepsis and control samples
The present study identified 158 metabolic pathways, 74 of which differed between sepsis patients and the controls (Figure 3A). The metabolite levels of all the lipid metabolic pathways, including those of arachidonic acid metabolism, steroid hormone biosynthesis, biosynthesis of unsaturated fatty acids, sphingolipid metabolism, fatty acid metabolism, fatty acid elongation, fatty acid degradation, and linoleic acid metabolism, were higher in the sepsis samples. Moreover, the metabolites related to the vast majority of the pathways (7/8) involving amino acid metabolism (including tyrosine metabolism, phenylalanine metabolism, histidine metabolism, phenylalanine, tyrosine and tryptophan biosynthesis, phosphonate and phosphinate metabolism, D-glutamine and D-glutamate metabolism, and cyano amino acid metabolism) were lower in the sepsis samples, whereas only those of tryptophan metabolism were increased. Compared to the control, all metabolites related to energy metabolism (including the gonadotropin-releasing hormone (GnRH) signaling pathway, ovarian steroidogenesis, and insulin secretion) and the vast majority (7/8) of those related to carbohydrate metabolism (including pentose phosphate pathway, glycolysis/gluconeogenesis, pyruvate metabolism, propanoate metabolism, butanoate metabolism, ascorbate and aldarate metabolism, and pentose and glucuronate interconversions) were significantly lower in the sepsis samples. The metabolic level of the hypoxia-inducible factor 1 (HIF-1) signaling pathway, which is one of the signal-transduction pathways regulated by oxygen availability, was lower in the sepsis samples (Figure 3B). In addition, 41 negative metabolites were found in living patients compared with deceased patients (Figure 3C).

Subgroups associated with sepsis-related organ dysfunction (Table 2)

Full table
A significant difference in phenylalanine levels was observed between the AKI and non-AKI groups. The present study identified seven metabolites that differed between these groups (Figure S1, Table S1). Furthermore, notable differences in the levels of three metabolites were observed between the ARDS and the non-ARDS groups, including 3-hydroxydecanoic acid, β-methylamino-l-alanine (BMAA), and 1H-imidazo[4,5-b]pyridine-2-carboxaldehyde (Figure S2, Table S2). Moreover, substantial differences in the levels of glutamine were observed between the SIMD and non-SIMD groups. In addition, seven metabolites also showed significant differences between these two subgroups. Only glutamine showed a positive correlation with the clinical indicator CK-MB (Figure S3, Table S3). A marked difference in phenylalanine levels was observed between the AHI and non-AHI groups. 3-methyl-2-oxopentanoic acid and 2-coumaranone were negatively correlated with total bilirubin (TBIL), while phenylalanine was positively correlated with TBIL (Figure S4, Table S4).
Discussion
Sepsis is defined as a life-threatening organ dysfunction caused by a dysregulated host response to infection (11). However, the specific factors that play key intermediary roles in sepsis remain unclear. Thus, elucidating the mechanisms underlying sepsis is essential to its diagnosis and therapy.
Cells are the basic units of tissues. Host cells play various biological roles, such as eliciting appropriate immune responses during sepsis. 5'-adenylate triphosphate (ATP) serves as the main source of cellular energy and is generated during aerobic metabolism (12). The pathophysiology of sepsis is relatively complex and involves interactions between the host and the microbial pathogens (13). Energy metabolism in sepsis is associated with an overall catabolic state leading to the breakdown of carbohydrates, lipids, and protein stores. Recent studies have revealed the importance of oxidative stress and mitochondrial dysfunction in sepsis. Metabolic changes associated with sepsis are orchestrated by interactions between immunoinflammatory and neuroendocrine stress responses (14). Therefore, the metabolic abnormalities caused by infection of the host are among the key elements of sepsis. In the present study, 124 metabolites showed differential levels between sepsis and control groups (P<0.05) (Figure 3A), which may reflect the metabolic condition during sepsis. Previous studies have shown that sepsis is often accompanied by significant metabolic changes, including metabolic hyperactivity, a high catabolic state, increased resting energy expenditure, increased protein and fat catabolism, negative nitrogen balance, insulin resistance, hyperglycemia, and increased liver glycogen production.
First, the amino acid metabolic process, which involves various molecules and biochemical metabolic pathways, is altered during sepsis. High-throughput MS indicated that six amino acids (tryptophan, glutamine, d-Proline, N-methoxycarbonyl-(octyl ester), and asparagine) exceeded its synthesis during sepsis, while the levels of seven amino acids (glutamic acid, cysteine, methionine, phenylalanine, putrescine, leucine, and aspartic acid) were elevated (Figure 3B). Among them, glutamine, leucine, and cysteine play important roles in sepsis (15). Studies have shown that the consumption of glutamine supplies carbon and nitrogen for cell proliferation through oxidization, and is converted into CO2 and pyruvate largely through the TCA cycle in mitochondria by glutaminolysis (16-19). Leucine is central to the regulation of global protein synthesis and the maintenance of cellular protein homeostasis by increasing mammalian target of rapamycin (mTOR) activity. The role of cysteine in the treatment of sepsis in improving survival rates has been confirmed in clinical and animal studies (20). We also found that the levels of amino acid derivatives were lower in sepsis patients compared to the healthy controls (Figure 3B), which indicates that the metabolism of proteins is a consumption process in sepsis.
Second, lipids are involved in the initiation and resolution of inflammation in sepsis. Oxidative stress affects the structure of polyunsaturated fatty acids in lipids by attacking their double bonds, and also initiates a chain reaction that generates peroxides and aldehydes that are involved in signaling and/or have deleterious consequences (21). In this study, we found that the levels of arachidonic acid, DHA, and EPA were lower in sepsis patients than in healthy controls (Figure 3B). The decrease in the oxidation rate of fatty acids is related to arachidonic acid, which is metabolized to prostaglandins and exerts mostly pro-inflammatory effects. In addition, anti-inflammatory and pro-resolving molecules are synthesized from EPA, DHA, and omega-3 fatty acids (22). Therefore, our findings serve as evidence for the above mechanism in sepsis.
Third, sepsis is characterized by a reduction in the function of the mitochondrial respiratory chain, coupled with free radical generation and bioenergetics reprograming toward glycolytic metabolism. The present study observed a decrease in the NADPH-NADP levels in sepsis patients (Figure 3B). It is understood that NADPH-oxidase and oxidative phosphorylation are activated and interact during a host response to infection. In addition, the increased levels of nitric oxide (NO) and superoxide (O2-) have been reported in sepsis patients, and are related to the prognosis outcomes. NO can modulate mitochondrial respiration and oxygen consumption through reversible binding and inhibition of complex IV, leading to the accumulation of NADPH and an increase in reactive oxygen species (ROS) production (23,24). The roles and mechanisms of other metabolites, including those of amino acid, fat, and sugar metabolism, which have been identified in this study, require further investigation.
The present study also analyzed metabolic pathways primarily using KEGG data. In our study, the metabolism of all lipids increased, while all energy and almost all carbohydrate metabolism decreased, indicating increased energy consumption. The metabolic HIF-1 signaling pathway exhibited differences between sepsis patients and healthy controls. HIF-1 is a major transcription factor that is involved in cellular adaptation to hypoxia. HIF-1α, one of the HIF-1 subunits, has been shown to modulate the expression of multiple target genes involved in angiogenesis, energy metabolism, apoptosis, autophagy, proliferation, and other adaptive responses to hypoxia. Recent studies have shown that during hypoxic conditions, HIF-1α migrates from the cytosol to the nucleus and then binds to its partner HIF-1β to form the HIF-1 complex. A decrease in the rate of the HIF-1 signaling pathway in sepsis patients results in lower HIF-1 accumulation rates (25-27). Our study illustrated that cells are in a state of hypoxia during sepsis (Figure S1), and thus, screening for metabolic pathways may help in the assessment of metabolic changes during sepsis.
All organs may be damaged during sepsis, and multiple organ dysfunction is a recognized complication. Further in-depth investigations of “life-threatening organ dysfunction” are essential to the management and treatment of sepsis. Paired subgroup analysis (i.e., AKI/non-AKI, ARDS/non-ARDS, SIMD/non-SIMD, and AHI/non-AHI) of plasma metabolites in sepsis patients has generated interesting information related to energy metabolism during sepsis.
The response that leads to sepsis-mediated AKI is complex and multifactorial, and involves interplay among inflammatory signaling, microcirculatory dysfunction, and alterations in cellular bioenergetics (28). The present study observed that increased levels of phenylalanine in the AKI group could predict the risk of kidney dysfunction during sepsis (Figure S1), and several studies have demonstrated the correlation with hydroxyl radical-induced tissue damage. The overproduction of hydroxyl radicals converts phenylalanine into para-, meta-, and ortho-tyrosine (p-, m-, and o-Tyr). We also detected a decrease in 2-coumaranone levels in the AKI group (Figure S1). 2-coumaranone shares some relevant similarities with hydrolyzed homocysteine thiolactone (HTL), which is considered a contributing factor to atherosclerosis (29). The decrease in 2-coumaranone levels may also be an intrinsic cause of excessively low vascular tone in sepsis-related AKI. However, the poor correlation between 2-coumaranone and carbamide, creatinine, uric acid, and KDIGO scores (Figure S1) indicates that 2-courmaranone may not be an appropriate biomarker for sepsis-complicated AKI. Furthermore, nonadecanoic acid levels are lower in sepsis patients with AKI (Figure S1). Nonadecanoic acid is a saturated fatty acid of bacteria [such as Escherichia coli (E. coli)] and thus may be a pathogenic factor, although the process of metabolism remains unclear (30).
Comparative analysis of ARDS/non-ARDS indicated that BMAA, which is present in various cyanobacterial species and certain terrestrial plants and seeds, may be a regionally specific phenomenon that is associated with environmental pollution and exposure. Another metabolite, 3-hydroxydecanoic acid (3-HD), is synthesized from glucose or fructose by recombinant E. coli harboring hydroxyacyl-ACP:CoA transacylase (phaG) and thioesterase II (tesB) or related compounds on the membrane of intact human erythrocytes (31,32). It appears to be related to pathogens or tissue damage and repair. Unfortunately, indicators related to “oxygen and index”, which are the key factors of ARDS, have not been found. Due to the complex etiology of sepsis, it is more difficult to analyze and screen metabolic markers in the ARDS subgroup.
Although the definition of SIMD remains controversial, we also attempted to filter the samples based on relevant criteria for subgroup analysis (i.e., SIMD/non-SIMD). We found that glutamine levels were lower in the non-SIMD group and higher in the SIMD group (Figure S3). In addition, we observed a correlation between glutamine levels and CK–MB, which is one of the key indices of myocardial damage. Microcirculatory alterations and mitochondrial damage, which reduce cellular energy production, have been considered a crucial mechanism of SIMD (33,34). Glutamine is the most abundant free amino acid in plasma and muscle tissue, and plays a major role in cell proliferation (35). It enters mitochondria primarily through the TCA cycle, which involves ammonia shuttling. The glutamine in plasma is transported into cells during sepsis, and SIMD may influence the metabolism of glutamine to glutamate in the mitochondria. Thus, a change in glutamine levels may provide clinical evidence for the energy metabolic mechanism underlying SIMD.
In addition, we observed statistical differences involving four metabolites [3-methyl-2-oxopentanoic acid, phenylalanine, 2-coumaranone, and benzeneacetic acid (3,4-dimethoxy-, methyl ester)] between the AHI and non-AHI subgroups (Figure S4). The aromatic amino acid phenylalanine is an essential substrate in humans. The demand for phenylalanine increases in the liver during infection. In addition, mitochondrial dysfunction causes tissue hypoxia and cell death, which may be the main reason for liver dysfunction secondary to sepsis. Modulators of mitochondrial activity (e.g., cytochrome C) are regulated by tyrosine phosphorylation (36). Thus, an increase in serum phenylalanine-tyrosine ratios may potentially be used in the screening for inflammatory diseases and in assessing the catabolic state of patients.
Some limitations of the present study should be noted. Firstly, we utilized a small study population. However, we matched the included sepsis patients with the controls, which could eliminate the interference caused by the small sample size to some extent. Furthermore, this paired analysis method is recognized in metabolomics research. Our research object was a sample group, and through the pairing of gender, age, and BMI, a standardized paired sample could be established. However, this pairing method also has certain limitations, as the population could have had genetic polymorphisms, host differences, and other issues. We have also attempted to reduce statistical errors using t-tests, which enabled us to realize the possibility of using small samples for metabolomics testing from our study. On the other hand, the difference in the etiology and pathogenesis of sepsis is currently the major constraint on in-depth studies of this syndrome. The present study utilized GC–MS to identify metabolites and metabolic pathways that are involved in sepsis and sepsis-associated organ dysfunction, which may serve as new clinical evidence for this specific condition. Therefore, focusing on the internal environment during sepsis may be a novel insight that could be utilized in sepsis management and therapy. This approach may also be applied to larger-scale studies. We are aware that some of the metabolites could not be recognized by our method and thus may influence our conclusions. Similarly, there is a possibility of omission in the subgroup analysis, suggesting that larger-scale studies are required. Ultimately, we aim to construct a metabolomics strategy to comprehensively characterize the sepsis metabolome that will drive biomarker discovery and/or identify target opportunities to improve the outcomes for sepsis patients.
Conclusions
GC–MS detection and metabolomic analysis of serum samples from sepsis patients identified some metabolites and metabolic pathways that are related to sepsis and sepsis-associated organ dysfunction. The three important metabolic pathways (energy metabolism, amino acid metabolism, and lipid metabolism) were downregulated in sepsis patients. This novel clinical evidence for the imbalance in the internal environment in energy metabolism disorders may elucidate the link between organ dysfunction and host response in sepsis. GC–MS analysis may also be potentially utilized as a detection tool for plasma metabolomics analysis of sepsis and related organ dysfunction.
Acknowledgments
Funding: This study was supported by Basic science and cutting-edge technology research projects of Chongqing Science and Technology Commission (cstc2016jcyjA0005 to FX, cstc2019jcyA0502 and cstc2020jcyj-msxmX1089 to SHL), Special fund of social undertakings and people’s livelihood guarantee of Chongqing science and technology commission (cstc2017shmsA130072, to FX), Medical research project of Chongqing City Health and Family Planning committee (2020GDRC001 to FX, 2020FYYX055 to CJW) and Chinese medicine science and technology project of Chongqing City Health and Family Planning committee (ZY201702071, to FX, 2019ZY3434 to SHL).
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-3562
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-3562
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-3562
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at: http://dx.doi.org/10.21037/atm-20-3562). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013), and was approved by the Ethics Committees of the First Affiliated Hospital of Chongqing Medical University (No. 2016-34). Informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jang JY, Lee SH, Shim H, et al. Serum oxygen radical activity and total antioxidation capacity are related with severities of surgical patient with sepsis: Prospective pilot study. J Crit Care 2017;39:131-6. [Crossref] [PubMed]
- Galley HF. Oxidative stress and mitochondrial dysfunction in sepsis. Br J Anaesth 2011;107:57-64. [Crossref] [PubMed]
- Dellinger RP, Levy MM, Rhodes A, et al. Surviving Sepsis Campaign: international guidelines for management of severe sepsis and septic shock, 2012. Intensive Care Med 2013;39:165-228. [Crossref] [PubMed]
- Ko ER, Yang WE, McClain MT, et al. What was old is new again: using the host response to diagnose infectious disease. Expert Rev Mol Diagn 2015;15:1143-58. [Crossref] [PubMed]
- Cheng SC, Scicluna BP, Arts RJ, et al. Broad defects in the energy metabolism of leukocytes underlie immunoparalysis in sepsis. Nat Immunol 2016;17:406-13. [Crossref] [PubMed]
- Zhang J, Hu ZD, Song J, et al. Diagnostic Value of Presepsin for Sepsis: A Systematic Review and Meta-Analysis. Medicine (Baltimore) 2015;94:e2158. [Crossref] [PubMed]
- Liu X, Ren H, Peng D. Sepsis biomarkers: an omics perspective. Front Med 2014;8:58-67. [Crossref] [PubMed]
- Tian H, Lam SM, Shui G. Metabolomics, a Powerful Tool for Agricultural Research. Int J Mol Sci 2016;17:1871. [Crossref] [PubMed]
- Jain A, Sankar J, Anubhuti A, et al. Prevalence and Outcome of Sepsis-induced Myocardial Dysfunction in Children with 'Sepsis' 'With' and 'Without Shock'-A Prospective Observational Study. J Trop Pediatr 2018;64:501-9. [Crossref] [PubMed]
- Levy MM, Fink MP, Marshall JC, et al. 2001 SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference. Crit Care Med 2003;31:1250-6. [Crossref] [PubMed]
- Rhodes A, Evans LE, Alhazzani W, et al. Surviving Sepsis Campaign: International Guidelines for Management of Sepsis and Septic Shock: 2016. Intensive Care Med 2017;43:304-77. [Crossref] [PubMed]
- Schmoch T, Uhle F, Siegler BH, et al. The Glyoxalase System and Methylglyoxal-Derived Carbonyl Stress in Sepsis: Glycotoxic Aspects of Sepsis Pathophysiology. Int J Mol Sci 2017;18:657. [Crossref] [PubMed]
- Ilia S, Briassoulis G. Sepsis is change and flows ever onwards. Expert Rev Anti Infect Ther 2017;15:515-7. [Crossref] [PubMed]
- Lukewich MK, Rogers RC, Lomax AE. Divergent neuroendocrine responses to localized and systemic inflammation. Semin Immunol 2014;26:402-8. [Crossref] [PubMed]
- Su L, Li H, Xie A, et al. Dynamic changes in amino acid concentration profiles in patients with sepsis. PLoS One 2015;10:e0121933. [Crossref] [PubMed]
- Tardito S, Oudin A, Ahmed SU, et al. Glutamine synthetase activity fuels nucleotide biosynthesis and supports growth of glutamine-restricted glioblastoma. Nat Cell Biol 2015;17:1556-68. [Crossref] [PubMed]
- Metallo CM, Vander Heiden MG. Metabolism strikes back: metabolic flux regulates cell signaling. Genes Dev 2010;24:2717-22. [Crossref] [PubMed]
- DeBerardinis RJ, Mancuso A, Daikhin E, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A 2007;104:19345-50. [Crossref] [PubMed]
- Newsholme EA, Crabtree B, Ardawi MS. Glutamine metabolism in lymphocytes: its biochemical, physiological and clinical importance. Q J Exp Physiol 1985;70:473-89. [Crossref] [PubMed]
- Tanaka KA, Kurihara S, Shibakusa T, et al. Cystine improves survival rates in a LPS-induced sepsis mouse model. Clin Nutr 2015;34:1159-65. [Crossref] [PubMed]
- Novak F, Borovska J, Vecka M, et al. Plasma Phospholipid Fatty Acid Profile is Altered in Both Septic and Non-Septic Critically Ill: A Correlation with Inflammatory Markers and Albumin. Lipids 2017;52:245-54. [Crossref] [PubMed]
- Buechler C, Pohl R, Aslanidis C. Pro-Resolving Molecules-New Approaches to Treat Sepsis? Int J Mol Sci. 2017;18:476. [Crossref] [PubMed]
- Nucci LA, Santos SS, Brunialti MK, et al. Expression of genes belonging to the interacting TLR cascades, NADPH-oxidase and mitochondrial oxidative phosphorylation in septic patients. PLoS One 2017;12:e0172024. [Crossref] [PubMed]
- Park DW, Zmijewski JW. Mitochondrial Dysfunction and Immune Cell Metabolism in Sepsis. Infect Chemother 2017;49:10-21. [Crossref] [PubMed]
- Mohamed AS, Hanafi NI, Sheikh Abdul Kadir SH, et al. Ursodeoxycholic acid protects cardiomyocytes against cobalt chloride induced hypoxia by regulating transcriptional mediator of cells stress hypoxia inducible factor 1alpha and p53 protein. Cell Biochem Funct 2017;35:453-63. [Crossref] [PubMed]
- Lv B, Hua T, Li F, et al. Hypoxia-inducible factor 1 alpha protects mesenchymal stem cells against oxygen-glucose deprivation-induced injury via autophagy induction and PI3K/AKT/mTOR signaling pathway. Am J Transl Res 2017;9:2492-9. [PubMed]
- Bar-Or D, Bar-Or R, Rael LT, et al. Oxidative stress in severe acute illness. Redox Biol 2015;4:340-5. [Crossref] [PubMed]
- Waltz P, Carchman E, Gomez H, et al. Sepsis results in an altered renal metabolic and osmolyte profile. J Surg Res 2016;202:8-12. [Crossref] [PubMed]
- Giacominelli-Stuffler R, Angelucci CB, Liberatoscioli L, et al. Relationships between paraoxon and 2-coumaranone hydrolytic activities in sera genotyped for PON1 Q192R polymorphism. Clin Biochem 2009;42:1512-6. [Crossref] [PubMed]
- Chiou RYY, Phillips RD, Zhao P, et al. Ethanol-Mediated Variations in Cellular Fatty Acid Composition and Protein Profiles of Two Genotypically Different Strains of Escherichia coli O157:H7. Applied and Environmental Microbiology 2004;70:2204-10. [Crossref] [PubMed]
- Zheng Z, Gong Q, Liu T, et al. Thioesterase II of Escherichia coli plays an important role in 3-hydroxydecanoic acid production. Appl Environ Microbiol 2004;70:3807-13. [Crossref] [PubMed]
- Kanaho Y, Sato T, Fujii T, et al. Shape-transforming action of myrmicacin (3-hydroxydecanoic acid) and some related compounds on the membrane of intact human erythrocytes. Chem Pharm Bull (Tokyo) 1981;29:3063-6. [Crossref] [PubMed]
- Neri M, Riezzo I, Pomara C, et al. Oxidative-Nitrosative Stress and Myocardial Dysfunctions in Sepsis: Evidence from the Literature and Postmortem Observations. Mediators Inflamm 2016;2016:3423450. [Crossref] [PubMed]
- Cimolai MC, Alvarez S, Bode C, et al. Mitochondrial Mechanisms in Septic Cardiomyopathy. Int J Mol Sci 2015;16:17763-78. [Crossref] [PubMed]
- Costa LC, Souza BN, Almeida FF, et al. Glutamine-Loaded Liposomes: Preliminary Investigation, Characterization, and Evaluation of Neutrophil Viability. AAPS PharmSciTech 2016;17:446-53. [Crossref] [PubMed]
- Moreno-Beltrán B, Guerra-Castellano A, Díaz-Quintana A, et al. Structural basis of mitochondrial dysfunction in response to cytochrome c phosphorylation at tyrosine 48. Proc Natl Acad Sci U S A 2017;114:E3041-50. [Crossref] [PubMed]
(English Language Editors: A. Kassem and J. Gray)


