T cell activation in utero: self or non-self discrimination
Fetal development in humans happens in the placenta, which has been believed to be a total sterile environment. As so, it should be quite unexpected to find activated T cells in cord blood, especially in the fetus blood.
Zhang et al. (1) published an article in Science Translational Medicine showing the presence of activated or memory T cells (TM) in human cord blood. Different from previous studies reporting the presence of CD45RO+ cells in cord blood (2), the authors elegantly used multicolor flow cytometry and transcriptomics to study cord blood activated T cells and their profile. It turned clear that cord blood TM from healthy individuals are pre-activated cells, capable of producing IFN-γ and IL-4 and express TBX1 and GATA3, as master genes for Th1 and Th2 development, respectively. Along with this line, the Th1 and Th2 cells also express chemokine receptors in accordance with their functional phenotype. Some of the cord blood T cells expressed RORc, although no IL-17A was detected unless the cells were stimulated ex-vivo in the presence of IL-23 and IL-1 eta. Also, by using transcriptomics, Zhang et al. (1) showed that the different TM subsets, based on their chemokine receptor expression, could be separated as a function of gene expression.
The important question raised by these findings is: how T cells are activated in a presumed sterile microenvironment? Are they responding to some unique non-inherited maternal antigen (NIMA)? Does a given specific antigen provide their stimulation? If so, the T cell repertoire should be skewed and a preferential representation of some specificity(ies) would be present. The authors compared the T cell repertoire of naïve (TN), regulatory (Treg) and memory cells from the collected healthy cord blood samples. Amazingly, the TM repertoire was polyclonal as well as the ones from TN and Treg, thus indicating no preferential expansion/stimulation of the TM subpopulation. Although being polyclonal, the repertoire differed between TN and TM but was quite similar when TM were compared with Treg. This suggests stimulation by autoantigens (Figure 1) since thymic derived Tregs are autorreative and have a repertoire similar to TM cells in the cord blood.
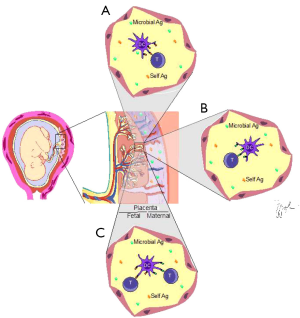
Another possibility to explain the generation of intra-uterus activated TM, although less likely, would be the presence of subclinical infections. However, the samples used were from delivery of healthy pregnant mothers without any sign of infection.
This led us to think if there are any extrinsic antigens in the placenta. In this respect, the presence of bacteria in the placenta has been associated with pre-term birth (3), as a polimicrobial disease. Placental contamination has been thought to happen from the outside of the mother’s body, with migration of the infectious agent arising from the vagina, into the uterine cavity. Curiously, the most frequent bacteria in the vagina of pregnant women are lactobacilli and this is not found in the baby, who harbors enterobacteria and bifidobacteria in their intestines, like their mom’s intestinal flora. How colonization happens is not clear, except that it is not coming from the external environment, but seems to be already present during pregnancy (4).
More recently, Aagaard et al. (5) published an article in Science Translational Medicine where placenta’s from 320 women were investigated regarding the presence of bacteria. These authors identified a microbial community in the placenta that correlated with antenatal infection, as expected. However, and most intriguing was the demonstration of a small, although consistent microbiome present in healthy pregnant women. Even more interesting, when they compared placental microbiome species composition with the microbiome of different body sites, placental microbiome is closer to that found in the oral cavity than to any other site of the body, including intestines. Yet, although being similar, placental and oral microbiomes are quite different when metabolism was evaluated, with overrepresentation of metabolism for cofactors and vitamins in the placentas than in any other site of the body, suggesting a metabolic role for these bacteria.
The fact that comes to light is that the placenta is not a sterile environment. On the contrary, it is a microenvironment rich in bacterial antigens and as so, being potentially able to stimulate adaptive immunity. Accordingly, we can expect the generation of co-stimulatory signals or the presence of pathogen associated molecular patterns (PAMPs) able to stimulate the innate immunity in order to trigger the adaptive immune response with resulting T cell activation (Figure 1). On the other hand, it is known that commensal microbiota can escape the immune response and whereas some species can be tolerogenic, others are not (6-10), contributing to inflammation or homeostasis.
If the activated T cells present in the cord blood of healthy normal babies, delivered by C-section or through vaginal route, are doing any good or not remains to be defined. On the same token, the microbiome might activate and be controlled by these few activated T cells, and this fine control might be important for intestinal maturation and maturation of the immune system.
In the near future we hope to understand the meaning of activated T cells during pregnancy. It will be necessary to establish the kinetics of appearance of such cells as well as the microbiome, and their mutual relationship. By now, the only thing we can say is that there are activated T cells in cord blood from healthy babies delivered from healthy mothers. If there is any meaning for these TM in the fetus/neonate and whether these are recognizing auto-antigens from the baby, maternal antigens or microbial antigens remain an interesting and important issue to be defined.
Acknowledgements
We are grateful to Rômulo Galvani for his help with the artwork. WS and AB are recipients of grants from Fiocruz, Faperj, CNPq (Brazil). WS is also granted by FOCEM (Mercosur).
Disclosure: The authors declare no conflict of interest.
References
- Zhang X, Mozeleski B, Lemoine S, et al. CD4 T cells with effector memory phenotype and function develop in the sterile environment of the fetus. Sci Transl Med 2014;6:238ra72.
- Theilgaard-Mönch K, Raaschou-Jensen K, Palm H, et al. Flow cytometric assessment of lymphocyte subsets, lymphoid progenitors, and hematopoietic stem cells in allogeneic stem cell grafts. Bone Marrow Transplant 2001;28:1073-82. [PubMed]
- Payne MS, Bayatibojakhi S. Exploring preterm birth as a polymicrobial disease: an overview of the uterine microbiome. Front Immunol 2014;5:595. [PubMed]
- Wassenaar TM, Panigrahi P. Is a foetus developing in a sterile environment? Lett Appl Microbiol 2014;59:572-9. [PubMed]
- Aagaard K, Ma J, Antony KM, et al. The placenta harbors a unique microbiome. Sci Transl Med 2014;6:237ra65.
- Mercadante AC, Perobelli SM, Alves AP, et al. Oral combined therapy with probiotics and alloantigen induces B cell-dependent long-lasting specific tolerance. J Immunol 2014;192:1928-37. [PubMed]
- Shan M, Gentile M, Yeiser JR, et al. Mucus enhances gut homeostasis and oral tolerance by delivering immunoregulatory signals. Science 2013;342:447-53. [PubMed]
- Kamada N, Seo SU, Chen GY, et al. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol 2013;13:321-35. [PubMed]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, et al. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 2004;118:229-41. [PubMed]
- Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 2009;9:313-23. [PubMed]


