
This article has an erratum available at: http://dx.doi.org/10.21037/atm-2022-12 the article has been update on 2022-07-14 at here.
SLC6A8 is involved in the progression of non-small cell lung cancer through the Notch signaling pathway
Introduction
Lung cancer is the most common cancer globally and the main cause of cancer-related mortality (1). Based on the World Health Organization classification, non-small cell lung cancer (NSCLC) accounts for approximately 85% of primary lung cancers, which are divided into adenocarcinoma, squamous cell carcinoma, and large cell carcinoma (2). Most patients with NSCLC are diagnosed at an advanced stage, leading to a very poor prognosis (3). Although considerable progress has been made in emerging technologies and newly developed targeted therapies in recent years, the 5-year overall survival rate for patients is only 19% (4-6).
The creatine transporter solute carrier family 6 member 8 (SLC6A8) is encoded by an 8.4-kb gene (7) and belongs to the subfamily of GABA transporters (8). SLC6A8 is a member of the sodium- and chloride-coupled transporter superfamily of neurotransmitters and organic permeants and is expressed in a variety of tissues, including brain, retina, skeletal muscle, and heart as well as a variety of epithelial cells (9). Heinbockel et al. reported that SLC6A8 is expressed in the bronchus, macrophages, and capillaries of chronic obstructive pulmonary disease (COPD) patients (10). SLC6A8 is known to participate in macrophage polarization and thus affects the immune response mediated by macrophages (11), whereas a lack of SLC6A8 can lead to arrhythmia or cardiomyopathy (12). Additionally, SLC6A8 has been shown to be related to aspects of the nervous system such as behavior (13), learning (14), and intelligence (15). Notably, many members of the GABA family have been confirmed to be involved in the occurrence and development of tumors (16-18). Loo et al. found that SLC6A8 can promote brain-type creatine kinase-mediated colon cancer metastasis (19). Further studies have shown that SLC6A8 expression is upregulated in stage III rectal cancer (20). However, the role and mechanism of SLC6A8 in the occurrence and development of NSCLC have not been studied.
We previously found via analysis of public databases [Oncomine, Gene Expression Omnibus (GEO), and The Cancer Genome Atlas (TCGA)] that SLC6A8 mRNA was significantly increased in lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD). Therefore, we speculated that SLC6A8 plays an important role in the occurrence and development of NSCLC. To confirm this hypothesis, we used the TCGA database to analyze the relationship between SLC6A8 gene expression and the prognosis of NSCLC. We then assessed the protein expression level of SLC6A8 in NSCLC clinical tissues and cell lines. Subsequently, we used in vitro cell transfection to study whether SLC6A8 is involved in the proliferation, migration, and invasion of NSCLC cells. Finally, using analysis of the gene set enrichment analysis (GSEA) data set and Western blotting, we determined that the expression of SLC6A8 is related to the Notch pathway and epithelial-mesenchymal transition (EMT). These results enable us to have a more comprehensive understanding of SLC6A8 in the field of oncology and to develop new strategies for targeted treatment of NSCLC. We present the following article in accordance with the MDAR reporting checklist (available at: http://dx.doi.org/10.21037/atm-20-5984).
Methods
Tissue collection
Thirty pairs of lung cancer tumor tissue and adjacent healthy tissue from patients at Qingdao Municipal Hospital were collected from January to July 2020. All patients had been diagnosed with NSCLC for the first time, and none of them had received radiotherapy or chemotherapy before surgery. The diagnosis of NSCLC was based on the 8th edition of the lung cancer metastasis classification recommended by the International Association for Lung Cancer Research staging committee. The select characteristics and clinical information, including sex, age, and tumor stage, were obtained from medical records. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Medical Ethics Committee of Qingdao Municipal Hospital affiliated to Qingdao University (No.: 2020yxy41) and informed consent was taken from all the patients. All specimens were anonymously processed in accordance with ethical and legal standards, and all methods were performed in accordance with relevant guidelines and regulations.
Cell culture and transfection
NSCLC cell lines, which included H1975 (CL-0298, Procell Life Science&Technology Co. Ltd., China), H520 (CL-0402, Procell Life Science&Technology Co. Ltd., China), H1299 (CL-0165, Procell Life Science&Technology Co. Ltd., China) and A549 (CL-0016, Procell Life Science&Technology Co. Ltd., China), were incubated in RPMI Medium 1640 (PM150110, Procell Life Science&Technology Co. Ltd., China) with 10% fetal bovine serum (10091-148, Gibco, USA) and 1% penicillin-streptomycin solution (15070-063, Gibco, USA) in a 5% CO2 incubator at 37 °C. The normal lung epithelial cell line BEAS-2B (CL-0496, Procell Life Science&Technology Co. Ltd., China) was cultured in Dulbecco’s Modified Eagle Medium (PM150210, Procell Life Science&Technology Co. Ltd., China) under the same conditions. Cells at a favorable growth state were passaged, frozen, and then recovered for the next experiment.
H1299 cells and H520 cells were cultured in 6-well plates. When the cells reached 70% confluence, SLC6A8 siRNA (GenePharma Co. Ltd., Shanghai, China) was transfected into H1299cells. SLC6A8 overexpression plasmid (G121621, YouBia, China) was transfected into H520 cells. SLC6A8 siRNA and SLC6A8 overexpression plasmid were separately mixed in Lipofectamine 2000 (#11668-019, Invitrogen, USA) and OPTI-MEM (#31985, Gibco, USA) according to the manufacturer’s instructions and then transfected into cells. Then added to the culture at 37 °C for 6 h. The culture medium was then replaced with complete medium. After 48 h, the cells were then harvested for functional analysis.
Bioinformatic analysis
We analyzed the RNA sequence data of the SLC6A8 gene in NSCLC tissue samples and normal lung tissue using the Oncomine database (http://www.oncoline.org), the TCGA database (https://cancergenome.nih.gov/), and NCBI/GEO/GSE101929 data. We used the TCGA database to analyze the correlation between SLC6A8 and matrix metalloproteinase-9 (MMP9). Online Kaplan-Meier analysis (http://kmplot.com/analysis/) was used to analyze the survival rate of 535 lung cancer patients with different expression levels of SLC6A8; GSEA-4.0.jar was used for GSEA) (https://www.gsea-msigdb.org/gsea/index.jsp).
Immunohistochemistry
Collected tissues were embedded in paraffin, sliced, deparaffinized, and then hydrated with ethanol, followed by antigen retrieval, and incubation with primary antibody anti-SLC6A8 (1:200, 20299-1-AP, Proteintech Group, Inc., China) at 4 °C overnight. The secondary antibody (1:50,000, BA1054, Boster, China) was developed for 20 minutes at 37 °C the next day. The slides were developed with DAB, and then counterstained and mounted; cells were observed and imaged under a microscope.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated using TRIzol reagent (15596-026, Ambion, USA), and reverse transcribed into cDNA using Hiscript Reverse Transcriptase (R101-01/02, Vazyme, China). We used SYBR-Green Master Mix (Q111-02, Vzyme, China) for amplification. The specific primers for SLC6A8 were as follows: forward 5'-GGTCAAATCCACGGGAAAGAT-3' and reverse 5'-CCACGAAGCCAGCAAAGAAGC-3'; primers for glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were as follows: forward 5'-TCAAGAAGGTGGTGAAGCAGG-3' and 5'-TCAAAGGTGGAGGAGTGGGT-3'. The relative mRNA expression of SLC6A8 was normalized to that of GAPDH and calculated using the 2−ΔΔCt method.
Western blotting
Cells cultured in the cell plate were washed with phosphate-buffered saline, lysed, and centrifuged at 4 °C to extract the supernatant. We used sodium dodecyl sulfate-polyacrylamide gel electrophoresis to separate proteins, which were then transferred to a polyvinylidene difluoride membrane (IPVH00010, Millipore, USA). The blots were blocked with 5% skim milk for 2 h and incubated with the primary antibody overnight at 4 °C. The primary antibodies used included anti-GAPDH (1:1,000, AB-P-R001, Hangzhou Xianzhi Biological Co. Ltd., China), anti-SLC6A8 (1:1,000, 20299-1-AP, Proteintech Group, Inc., China), anti-N-cadherin (1:1,000, Ab124397, Abcam, UK), anti-E-cadherin (1:5,000, 20874-1-AP, Proteintech Group, Inc., China), anti-MMP9 (1:1,000, Ab76003, Abcam, UK), anti-Notch-1 (1:1,000, Ab52627, Abcam, UK), and anti-Notch-2 (1:1,000, 28580-1-AP, Proteintech Group, Inc., China) antibodies. The membranes were washed the next day and incubated with HRP-labeled secondary antibody at 37 °C for 2 h. The secondary antibodies were HRP-labeled goat anti-rabbit (1:50,000, BA1054, Boster, China) and goat anti-mouse (1:50,000, BA1051, Boster, China) secondary antibodies. Finally, we developed the signal by pressing the tablet and analyzed with BandScan.
CCK8 test
After transfection and culture in a 96-well plate, 10 µL CCK8 (C0037, Beyotime, China) was added to each well, and the plate incubated at 37 °C for 4 h. The absorbance value (OD at 450 nm) of each well was then measured with a microplate reader (MULTISKAN MK3, Thermo, USA).
5-ethynyl-2'-deoxyuridine (EdU) text
The EdU assay (C0081s, Beyotime, China) was used to determine cell proliferation. EdU labelling, cell immobilization, Apollo staining and DNA staining were used ac-cording to the instructions, and finally, images were acquired and analysed by fluorescence microscopy.
Cell cycle detection
After transfection in a 6-well plate, the cells were collected and centrifuged. The resuspended cells were fixed with ethanol at 4 °C for more than 4 h. The cells were resuspended by centrifugation and analyzed using a cell cycle detection kit (KGA512, Nanjing KGI Biotechnology Biological Development Co. Ltd., China). Finally, flow cytometry (FACSCalibur, BD Biosciences, USA) was used to determine the percentage of G1-, S-, and G2-phase cells to the total cell number.
Cell scratch test
The cells were transfected in a 6-well plate and trypsinized after culture. The cell density was adjusted to 90% and the cells were re-seeded in a 6-well plate and cultured overnight in 5% CO2 saturated humidity at 37 °C. When the cells covered the bottom of the 6-well plate, a pipette tip was used to scratch the cell layer. The cells were then washed with phosphate-buffered saline, and serum-free medium was added; images of the cell layer were taken at 0 and 24 h using a microscope (IX51, Olympus, Japan).
Cell invasion test
Medium (1640 plus 10% FBS) pre-cooled to 4 °C was added to a Transwell chamber. Matrigel (corning, USA) was added to the upper chamber and incubated at 37 °C for 4–5 h to dry into a gel. Finally, the cell suspension of each group was seeded into the upper chamber of the Transwell and cultured for 24 h. We removed the Transwell chamber and fixed the cells with an ice-cold ethanol solution for 1 h. The cells were then finally stained and washed and images were taken under a microscope (Olympus, Japan).
Statistical analysis
Statistical analysis was evaluated using SPSS 24.0 (Chicago, IL, USA). The Kaplan-Meier curve was used to calculate the survival curve and survival rate. The correlation analysis was conducted with Pearson analysis, and the comparison between groups was conducted with Student’s t-test or one-way analysis of variance. Data are expressed as mean ± standard deviation; a P value <0.05 was considered as statistically significant. All independent experiments are performed at least thrice.
Results
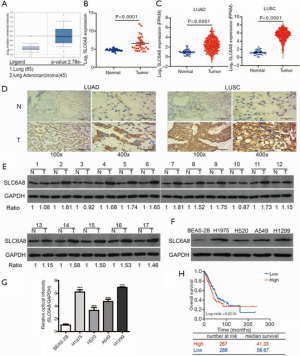
SLC6A8 is upregulated in NSCLC
To explore the mechanism underlying the effects of SLC6A8 in NSCLC, we analyzed the expression level of SLC6A8 in NSCLC and normal lung tissues using several tumor databases (Oncomine, GEO, and TCGA). The mRNA expression of SLC6A8 was significantly higher in tumor tissue than in the corresponding healthy tissue (Figure 1A,B,C). Immunohistochemistry and Western blotting results showed that the expression level of SLC6A8 in tumor tissues was higher compared with that in adjacent lung tissues (Figure 1D,E). When compared with that in the normal lung epithelial cell line BEAS-2B, SLC6A8 expression was consistently and significantly increased in all NSCLC cell lines (H1975, H520, A549, and H1299) (Figure 1F,G). These results indicate that SLC6A8 is upregulated in NSCLC cells and tissues. Finally, we used the public database Kaplan-Meier plotter to analyze the relationship between the expression of SLC6A8 and the prognosis of NSCLC patients to demonstrate that the prognosis of lung cancer patients was poorer when there was higher expression of SLC6A8 (Figure 1H). We analyzed the median survival time of SLC6A8 using the TCGA database. The results showed that the median survival time of the higher expression of SLC6A8 was 41.33 months, while the lower group was 56.67 months (Figure 1H). This proves that there is a significant difference of survival between NSCLC with higher SLC6A8 and those with lower SLC6A8.
SLC6A8 promotes the proliferation of NSCLC cells in vitro
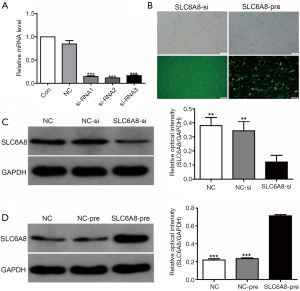
SLC6A8 expression was highest in the NSCLC cell line H1299 cell, whereas that in H520 cells was relatively low (Figure 1F,G); therefore, we induced SLC6A8 silencing (SLC6A8-si) in H1299 cells and overexpression (SLC6A8-pre) in H520 cells to study the functionality of SLC6A8. SLC6A8-si and SLC6A8-pre significantly downregulated or upregulated the expression of SLC6A8 protein, respectively, indicating that the cell model was successfully constructed (Figure 2A,B,C,D).
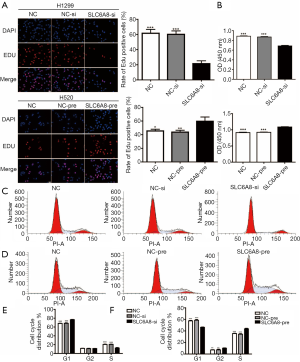
EdU assays and CCK8 assays showed that knockdown of SLC6A8 significantly inhibited the proliferation of H1299 cells (Figure 3A,B), whereas overexpression of SLC6A8 significantly promoted the proliferation of H520 cells (Figure 3A,B). Cell cycle analysis (Figure 3C,D,E,F) showed that downregulation of SLC6A8 increased the proportion of H1299 cells in G1 phase and decreased the proportion of cells in S phase (Figure 3C,E); however, upregulation of SLC6A8 reduced the proportion of H520 in G1 phase and increased the proportion of cells in S phase (Figure 3D,F). Thus, SLC6A8 can promote the proliferation of lung cancer cells by affecting the cell cycle distribution of NSCLC.
SLC6A8 promotes the migration and invasion of NSCLC cells in vitro
To determine the effect of SLC6A8 on the migration and invasion of NSCLC cells, we conducted a scratch test and a Transwell invasion test. The results showed that compared with the control group, the migration and invasion abilities of H1299 cells in the SLC6A8-si group were significantly reduced (Figure 4A,B,C). In contrast, those of H520 cells in the SLC6A8-pre group were significantly higher than those seen in the control group (Figure 4A,B,C).
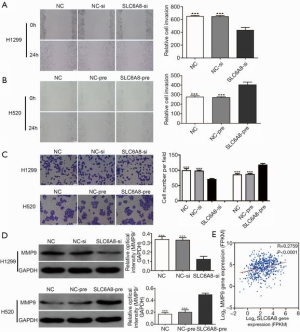
MMPs, including MMP9, can degrade and modify many of the components of the extracellular matrix and basement membrane and play an important role in cancer invasion and migration (21,22). Western blotting showed that knockdown of SLC6A8 expression downregulated the expression of MMP9, whereas overexpression of SLC6A8 clearly upregulated the expression of MMP9 (Figure 4D). Subsequently, we used the TCGA data set to analyze the correlation between SLC6A8 and MMP9; we observed that the expressions of SLC6A8 and MMP9 were positively correlated (Figure 4E). These results suggest that SLC6A8 is involved in the invasion and migration of NSCLC cells.
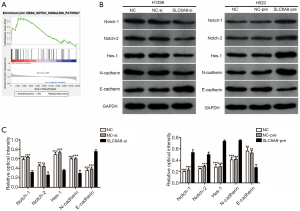
SLC6A8 mediates EMT to promote NSCLC progression through the Notch signaling pathway
At present, the specific regulatory mechanism of SLC6A8 in NSCLC is unclear. We used GSEA software to explore SLC6A8-related signal pathways. Analysis of the GSEA data set showed that SLC6A8 may promote the progression of NSCLC through the Notch signaling pathway (Figure 5A). Furthermore, Western blotting indicated that the expressions of Notch1, Notch2 and Hes1 in the SLC6A8 knockdown group were significantly reduced compared with those of control (Figure 5B), although they were significantly increased in the SLC6A8 overexpression group (Figure 5B). Studies have reported that Notch signaling pathway can promote the progression of NSCLC by enhancing EMT (23). Subsequently, we used Western blotting to detect the expression of EMT-related proteins (E-cadherin and N-cadherin) in cells where SLC6A8 was overexpressed or silenced. The results showed that SLC6A8-si significantly increased the expression of E-cadherin and decreased the expression of N-cadherin in NSCLC cells (Figure 5B,C). In contrast, the expressions of E-cadherin and N-cadherin in the SLC6A8-pre group were significantly lower and higher than that in the control group, respectively (Figure 5B,C).
Discussion
Lung cancer is the most prevalent cancer globally, and there has been extensive research on the pathogenesis of and therapeutic targets for this disease. Here, we explored the role of SLC6A8 in NSCLC. We found that the expression of SLC6A8 was increased in NSCLC and that this was related to the poor prognosis of patients. In addition, overexpression of SLC6A8 in NSCLC cell lines activated the Notch signaling pathway to promote NSCLC proliferation and metastasis, whereas downregulation of SLC6A8 suppressed the above effects. Therefore, this study shows for the first time that SLC6A8 plays a role in the development of NSCLC.
The SLC family includes several membrane transporters that are abnormally expressed in a variety of tumors and can participate in tumor pathogenesis (16-18). SLC6A8 has recently been found to be a new cancer-promoting gene. Loo et al. reported that SLC6A8 expression is upregulated in patients with advanced colon cancer and that this is closely related to tumor metastasis (19,20). However, the role of SLC6A8 in lung cancer remains unclear. In this study, we initially analyzed public databases and determined that the expression of SLC6A8 mRNA was much higher than that present in corresponding healthy tissues. In addition, immunohistochemical and Western blot analysis showed that the expression of SLC6A8 in lung cancer tissues was clearly higher than that in adjacent tissues. In vitro, the overexpression or knockdown of SLC6A8 can respectively enhance or inhibit the proliferation, migration, and invasion of NSCLC cells. More importantly, bioinformatic analysis showed that the prognosis of NSCLC patients was poorer in the SLC6A8 mRNA high expression group than in the low expression group. Therefore, we speculated that SLC6A8 is involved in the occurrence and development of NSCLC, although the underlying molecular mechanism was unclear.
Notch receptor is a transmembrane protein composed of a series of different protein modules that relies on cell contact to regulate the response of the body to environmental signals (24). The Notch receptor has been shown to play an important role in the progression of cancers of the lung, colon, liver, and gastrointestinal tract as well as of other tumors (25-29). Studies have found that Notch1/2 promotes the invasion and metastasis of NSCLC by activating EMT, which is related to the poor prognosis of patients (23). Sharif et al. found that Notch1/2 is upregulated in NSCLC and promotes the proliferation of NSCLC cells (29). However, the relationship between SLC6A8 and Notch has not been studied. To clarify the relationship between these proteins, we conducted a gene set pathway enrichment analysis and found that the Notch signaling pathway was significantly enriched in the SLC6A8 high expression group. Further experiments found that overexpression of SLC6A8 can promote the expression of Notch1, Notch2, and N-cadherin (EMT marker molecule) in NSCLC cells. Knockdown of SLC6A8 can inhibit the above effects. In summary, we know that SLC6A8 may promote the proliferation and invasion of NSCLC cells by regulating the Notch signaling pathway.
This research has several limitations. First, this study initially explored the role of SLC6A8 in NSCLC, but this has not been studied in vivo, which we will address in subsequent studies. Second, although the regulatory relationship of SLC6A8/Notch has been studied in vitro, this has not been verified in vivo and will be assessed in subsequent research.
Conclusions
SLC6A8 participates in the proliferation and metastasis of NSCLC, which is related to the poor prognosis of NSCLC. Therefore, SLC6A8 is expected to become a new prognostic predictor and therapeutic target for NSCLC.
Acknowledgments
We would like to thank Prof. Johnnie for his help in polishing the language of our paper.
Funding: This work was supported by the Development of Class A Key Disciplines by Qingdao Healthy Committee.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-5984
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-5984
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-5984). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of Qingdao Municipal Hospital Affiliated to Qingdao University (No. 2020yxy41) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Oser MG, Niederst MJ, Sequist LV, et al. Transformation from non-small-cell lung cancer to small-cell lung cancer: molecular drivers and cells of origin. Lancet Oncol 2015;16:e165-72. [Crossref] [PubMed]
- Ramalingam SS, Owonikoko TK, Khuri FR. Lung cancer: New biological insights and recent therapeutic advances. CA Cancer J Clin 2011;61:91-112. [Crossref] [PubMed]
- Rosell R, Karachaliou N. Lung cancer in 2014: optimizing lung cancer treatment approaches. Nat Rev Clin Oncol 2015;12:75-6. [Crossref] [PubMed]
- Cortes-Dericks L, Galetta D. The therapeutic potential of mesenchymal stem cells in lung cancer: benefits, risks and challenges. Cell Oncol (Dordr) 2019;42:727-38. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Rosenberg EH, Almeida LS, Kleefstra T, et al. High prevalence of SLC6A8 deficiency in X-linked mental retardation. Am J Hum Genet 2004;75:97-105. [Crossref] [PubMed]
- Colas C, Banci G, Martini R, et al. Studies of structural determinants of substrate binding in the Creatine Transporter (CreaT, SLC6A8) using molecular models. Sci Rep 2020;10:6241. [Crossref] [PubMed]
- Fezai M, Jemaa M, Fakhri H, et al. Down-Regulation of the Na+,Cl- Coupled Creatine Transporter CreaT (SLC6A8) by Glycogen Synthase Kinase GSK3ss. Cell Physiol Biochem 2016;40:1231-8. [Crossref] [PubMed]
- Heinbockel L, Marwitz S, Schromm AB, et al. Identification of novel target genes in human lung tissue involved in chronic obstructive pulmonary disease. Int J Chron Obstruct Pulmon Dis 2018;13:2255-9. [Crossref] [PubMed]
- Ji L, Zhao X, Zhang B, et al. Slc6a8-Mediated Creatine Uptake and Accumulation Reprogram Macrophage Polarization via Regulating Cytokine Responses. Immunity 2019;51:272-284.e7. [Crossref] [PubMed]
- Anselm IA, Coulter DL, Darras BT. Cardiac manifestations in a child with a novel mutation in creatine transporter gene SLC6A8. Neurology 2008;70:1642-4. [Crossref] [PubMed]
- Griswold AJ, Ma D, Cukier HN, et al. Evaluation of copy number variations reveals novel candidate genes in autism spectrum disorder-associated pathways. Hum Mol Genet 2012;21:3513-23. [Crossref] [PubMed]
- Hahn KA, Salomons GS, Tackels-Horne D, et al. X-linked mental retardation with seizures and carrier manifestations is caused by a mutation in the creatine-transporter gene (SLC6A8) located in Xq28. Am J Hum Genet 2002;70:1349-56. [Crossref] [PubMed]
- van de Kamp JM, Betsalel OT, Mercimek-Mahmutoglu S, et al. Phenotype and genotype in 101 males with X-linked creatine transporter deficiency. J Med Genet 2013;50:463-72. [Crossref] [PubMed]
- Chen C, Cai Z, Zhuo Y, et al. Overexpression of SLC6A1 associates with drug resistance and poor prognosis in prostate cancer. BMC Cancer 2020;20:289. [Crossref] [PubMed]
- Sung HY, Yang SD, Park AK, et al. Aberrant Hypomethylation of Solute Carrier Family 6 Member 12 Promoter Induces Metastasis of Ovarian Cancer. Yonsei Med J 2017;58:27-34. [Crossref] [PubMed]
- Tran TT, Mu A, Adachi Y, et al. Neurotransmitter transporter family including SLC6A6 and SLC6A13 contributes to the 5-aminolevulinic acid (ALA)-induced accumulation of protoporphyrin IX and photodamage, through uptake of ALA by cancerous cells. Photochem Photobiol 2014;90:1136-43. [Crossref] [PubMed]
- Loo JM, Scherl A, Nguyen A, et al. Extracellular metabolic energetics can promote cancer progression. Cell 2015;160:393-406. [Crossref] [PubMed]
- Katkoori VR, Shanmugam C, Jia X, et al. Prognostic significance and gene expression profiles of p53 mutations in microsatellite-stable stage III colorectal adenocarcinomas. PLoS One 2012;7:e30020. [Crossref] [PubMed]
- Zuo J, Wen M, Li S, et al. Overexpression of CXCR4 promotes invasion and migration of non-small cell lung cancer via EGFR and MMP-9. Oncol Lett 2017;14:7513-21. [Crossref] [PubMed]
- Li W, Jia MX, Wang JH, et al. Association of MMP9-1562C/T and MMP13-77A/G Polymorphisms with Non-Small Cell Lung Cancer in Southern Chinese Population. Biomolecules 2019;9:107. [Crossref] [PubMed]
- Liu D, Lin L, Wang Y, et al. PNO1, which is negatively regulated by miR-340-5p, promotes lung adenocarcinoma progression through Notch signaling pathway. Oncogenesis 2020;9:58. [Crossref] [PubMed]
- Aster JC, Pear WS, Blacklow SC. The Varied Roles of Notch in Cancer. Annu Rev Pathol 2017;12:245-75. [Crossref] [PubMed]
- Jiang J, Ren H, Xu Y, et al. TRIM67 Promotes the Proliferation, Migration, and Invasion of Non-Small-Cell Lung Cancer by Positively Regulating the Notch Pathway. J Cancer 2020;11:1240-9. [Crossref] [PubMed]
- Jin L, Vu T, Yuan G, et al. STRAP Promotes Stemness of Human Colorectal Cancer via Epigenetic Regulation of the NOTCH Pathway. Cancer Res 2017;77:5464-78. [Crossref] [PubMed]
- Li H, Lan T, Xu L, et al. NCSTN promotes hepatocellular carcinoma cell growth and metastasis via beta-catenin activation in a Notch1/AKT dependent manner. J Exp Clin Cancer Res 2020;39:128. [Crossref] [PubMed]
- Kang HG, Kim WJ, Noh MG, et al. SPON2 Is Upregulated through Notch Signaling Pathway and Promotes Tumor Progression in Gastric Cancer. Cancers (Basel) 2020;12:1439. [Crossref] [PubMed]
- Sharif A, Shaji A, Chammaa M, et al. Notch Transduction in Non-Small Cell Lung Cancer. Int J Mol Sci 2020;21:5691. [Crossref] [PubMed]


