
Late response to rosuvastatin and statin-related myalgia due to SLCO1B1, SLCO1B3, ABCB11, and CYP3A5 variants in a patient with Familial Hypercholesterolemia: a case report
Introduction
Familial hypercholesterolemia (FH) is a genetic metabolic disease that leads to increased high low-density lipoprotein (LDL) cholesterol, which is a risk factor for early atherosclerosis and cardiovascular diseases (1). FH is usually treated with high-dose statins, which are inhibitors of 3-hydroxy-3-methyl-glutaryl coenzyme A reductase (HMGR), a key enzyme in cholesterol biosynthesis pathway. Rosuvastatin is one of the most effective statins, probably due its hydrophilicity, that confers selectivity to hepatic cells, higher affinity to HMGR, and lower rates of statin-related adverse events (SRAE) compared to other statins. It is poorly metabolized by CYP2C9 and CYP2C19, while 72% of the non-metabolized molecules are excreted via biliary system. Therefore, rosuvastatin blood levels rely on the activity of membrane transporters, mainly of solute carrier (SLC) and ATP-binding cassette (ABC) families, highly expressed in intestine, liver, and kidney (2).
Pharmacogenetic studies have shown that loss-of-function variants in genes encoding OATPs, such as SLCO1B1, SLCO2B1, and SLCO1B3, and ABCs have been associated with variability in low-density lipoprotein cholesterol (LDL-c) reduction and higher risk of SRAE (3). The importance of considering the combined effect of variants in key genes for pharmacogenetic analyses has been increasingly evident (4). In this case report, we discuss how variants in genes participating in different stages of statin pharmacokinetics pathway possibly affected the time to response to rosuvastatin and the risk of SRAE in a female FH patient. To the best of our knowledge, this is the first report of a pharmacogenetic analysis on a case of late rosuvastatin response. This case is reported in accordance with the CARE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-5540).
Case presentation
A 26-year-old Caucasian female patient with definite diagnosis of FH according to Dutch Lipid Clinic Network MEDPED criteria (5) was invited to participate in an intervention study in June 2019. She was previously included in a FH sequencing study (May 2018), in which a panel of 84 genes involved in lipid homeostasis and drug metabolism was sequenced using exon-targeted gene sequencing (NGS). All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committees and with the Helsinki Declaration (as revised in 2013). Written informed consent was obtained from the patient for both studies.
The patient carries the variant LDLR rs28941776 (c.1646G>A, p.Gly549Asp), which has been associated with FH and is classified as pathogenic according to the American College of Medical Genetics (ACMG) guidelines (6).
Her clinical history included high levels of total cholesterol and LDL-c since childhood. In 2008, at the age of 15 years, she had an abnormal lipid profile even under a daily treatment with simvastatin 10 mg and ezetimibe 10 mg. Laboratory analyses showed a total cholesterol of 324 mg/dL, LDL-c 264 mg/dL, high-density lipoprotein cholesterol (HDL-c) 46 mg/dL, and triglycerides 71 mg/dL. In 2014, she was diagnosed with hypothyroidism and treated with levothyroxine 25 µg/day, which was gradually increased to 100 µg/day in 2019. She also had a pregnancy history in January 2017.
Her therapy history included simvastatin, which led to severe myopathy in 2008, with marked increase in serum creatine kinase (CK) to 1,080 U/L (4.7-fold the upper reference value). The cholesterol-lowering therapy was changed to pravastatin 20 mg and ezetimibe 10 mg daily until May 2011, when she reported another episode of myalgia. Pravastatin was withdrawn and atorvastatin 20 mg was introduced, also associated with ezetimibe 10 mg. Three months later, in August 2011, she reported interrupting atorvastatin treatment due to myalgia. Rosuvastatin 10 mg was then introduced, also associated with ezetimibe 10 mg, after which she showed an LDL-c level of 125 mg/dL and never reported myalgia again. However, her lipid profile worsened throughout the years even under rosuvastatin treatment, with her LDL-c reaching 194 mg/dL with rosuvastatin 20 mg.
The patient had no history of liver or kidney impartment, HIV, coronary artery disease (CAD), diabetes, obesity, cardiovascular events, and did not smoke or drink. Her mother and grandmother had a history of FH, but not CAD or cardiovascular events, while her father had hypertension and type 2 diabetes.
In the intervention study, the patient was seen four times (V1 to V4) in 5 months, and clinical history and therapy data were obtained. The protocol consisted of a 6-week rosuvastatin wash-out period, after which rosuvastatin was reintroduced for additional 6 weeks, when treatment response was evaluated. Adherence to treatment was assessed in each timepoint using the translated and validated version of the Brief Medication Questionnaire (BMQ) (7) and blood samples were taken in each visit for laboratory testing.
The lipid profile during the follow-up is shown in Figure 1. In April 2019 (V1), the patient was taking rosuvastatin 40 mg, ezetimibe 10 mg, and levothyroxine 88 µg daily. She reported experiencing muscle pain after recently increasing rosuvastatin dose from 20 to 40 mg/day. Her lipid profile was altered (total cholesterol 376 mg/dL, LDL-c 263 mg/dL, HDL-c 67 mg/dL, triglycerides 234 mg/dL) without increase in CK levels. She reported being active, running 2 km 2–3 times a week, and had a healthy diet, eating more than five portions of vegetables daily. Her TSH and T4 levels were normal. Rosuvastatin 40 mg was then discontinued for wash-out, ezetimibe was maintained, and levothyroxine dose was increased to 100 µg/day.
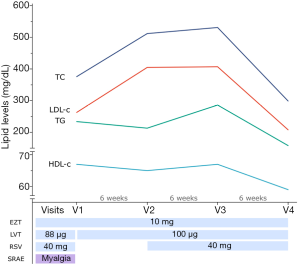
In June 2019 (V2), after undergoing a 6-week rosuvastatin wash-out period between V1 and V2, her lipid profile worsened (total cholesterol 512 mg/dL, LDL-c 405 mg/dL, HDL-c 65 mg/dL, triglycerides 213 mg/dL). Because the patient reported myalgia in V1 (rosuvastatin 40 mg), the physician prescribed rosuvastatin 20 mg/day for six weeks. Surprisingly, in August 2019 (V3), the lipid profile (total cholesterol 531 mg/dL, LDL-c 407 mg/dL, HDL-c 67 mg/dL, triglycerides 286 mg/dL) did not change compared to V2. The patient reported experiencing no myalgia to rosuvastatin 20 mg. In September 2019 (V4), her lipid profile improved (total cholesterol 299 mg/dL, LDL-c 208 mg/dL, HDL-c 59 mg/dL, triglycerides 158 mg/dL) and she continued not experiencing myalgia to rosuvastatin.
During the follow-up period, serum TSH and T4 levels remained unchanged, suggesting that her hypothyroidism was controlled and did not influence the lipid profile. Moreover, serum CK did not show any abnormality, which indicates no muscle damage due to statin treatment.
The patient also reported being adherent to treatment. In the BMQ adherence questionnaire, she reported forgetting the lipid-lowering medications 2 days in the week before V1 (71.4% adherence) and 1 day in the week before V3 (85.7% adherence).
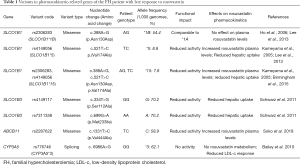
The genetic profile of the patient is shown in Table 1. She carries five missense variants in SLCO1B1, SLCO1B3, and ABCB11. She is also homozygote for the CYP3A5*3 (rs776746) splicing variant. No other missense variants described as impacting rosuvastatin response were found in CYP3A4, CYP2C9, CYP2C19, or other drug transporters, such as ABCG2 (data not shown).
Full table
Discussion
In heterozygous FH patients, LDL-c level reductions of 47.1% have been observed after a 6-week treatment with rosuvastatin 20 mg (8). The patient, however, did not experience any changes in LDL-c levels at week 6 (V3) of rosuvastatin 20 mg treatment, with a 48.9% LDL-c reduction only at week 12 (V4) of therapy.
The delayed rosuvastatin response could be explained by modifications in the therapy scheme during the follow-up period. However, the only change was in levothyroxine dose, that was increased from 88 to 100 µg in V1. It is unlikely that the late response is due to an adaptation to the new levothyroxine dose. The patient was already on treatment with levothyroxine 88 µg before V1; moreover, changes in cholesterol due to an adaptation period should be reflected in her lipid profile in V3, not only in V4. Another possible explanation is a lack of adherence from V2 to V3; however, the patient showed a similar treatment adherence in V3 and V1, which should lead to a similar lipid profile between visits. Furthermore, drug interactions between rosuvastatin, levothyroxine, and ezetimibe that could affect treatment response were not detected, excluding this possibility.
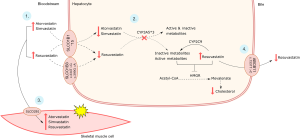
Pharmacokinetics-related genes may have contributed to the late response to rosuvastatin (Figure 2). The patient carries two variants in SLCO1B1, c.388A>G (SLCO1B1*1B) and c.521T>C (SLCO1B1*5), that are important determinants of rosuvastatin response. SLCO1B1*5 is a loss-of-function variant that decreases the hepatic uptake and increases blood levels of statins (9) (Table 1). SLCO1B1*1B has shown comparable activity to the functional *1A variant in in vitro functional studies (10). SLCO1B1*1B and *5 variants are in linkage disequilibrium (LD) and form the SLCO1B1*15 haplotype, that also reduced rosuvastatin uptake in functional studies with HEK293 and HeLa cells (11). The decreased liver uptake caused by these SLCO1B1 variants has been associated with increased plasma levels of rosuvastatin in pharmacokinetics studies (9) (Table 1).
SLCO1B3 is also an important gene that encodes an influx transporter for rosuvastatin. The patient was homozygous for both SLCO1B3 c.334T>G and c.699G>A, which are in strong LD (12). In an in vitro study, HeLa cells transfected with SLCO1B3 c.334G and c.699A haplotype showed a 13% decrease in rosuvastatin uptake, while for other substrates, such as cholecystokinin-8, an even more marked decrease of 57% was observed (13) (Table 1).
Although the effect of SLCO1B3 c.334G and c.699A haplotype in rosuvastatin uptake is not sufficient to explain the delayed response, it might be significant when combined with the effect of the decreased function haplotype SLCO1B1*15. While SLCO1B1*5 and SLCO1B1*15 are associated with higher plasma levels of rosuvastatin, previous studies failed to find an association between these variants and LDL-c reduction in response to short- and long-term rosuvastatin treatments (9). Therefore, the simultaneous presence of decreased function SLCO1B1 and SLCO1B3 haplotypes possibly caused a marked reduction of rosuvastatin intrahepatic concentration, resulting in the lack of response observed in V3.
ABCB11 encodes the efflux protein ABCB11, which plays an important role in rosuvastatin bile excretion. In a recent study, ABCB11 c.1331C allele has been associated to increased plasma rosuvastatin levels in healthy subjects (14) (Table 1). This variant possibly causes lower rosuvastatin excretion via bile, which in turn would increase intrahepatic rosuvastatin concentrations. Therefore, this mechanism could explain why even in the presence of low function SLC variants, the patient showed a late but evident LDL-c reduction after 12 weeks of rosuvastatin treatment.
The patient also carries the homozygous form of CYP3A5*3, an intronic variant that results in undetectable expression of CYP3A5 (15). The GEOSTAT-1 study reported that dyslipidemic patients carrying CYP3A5*3/*3 had lower LDL-c reduction after three-month rosuvastatin 10 mg treatment compared to carriers of *1/*1 or *1/*3 (Table 1). It was suggested that the metabolite produced by CYP3A5 also plays a role in HMGR inhibition, potentiating the response to rosuvastatin, which is why CYP3A5 non-expressors have reduced LDL-c response to rosuvastatin (16). CYP3A5*3 possibly impaired the patient’s response time to rosuvastatin, but in lower extent, as CYP3A5 does not participate markedly in rosuvastatin metabolism.
In addition to the delayed response to rosuvastatin, the patient experienced myalgia associated with rosuvastatin 40 mg/day and other statins, as previously commented. This SRAE may be due to SLCO1B1 variants. SLCO1B1*5 and SLCO1B1*15 have been extensively associated with myopathy to simvastatin. A systematic review and meta-analysis reported that carriers of the C allele of SLCO1B1*5 (c.521T>C) showed a higher risk of myotoxicity (17). Additionally, SLCO1B1*5 has been associated to rosuvastatin myotoxicity in previous studies (18,19). It has been suggested that it causes higher efflux of statins, increasing statin exposure and, therefore, the risk of myalgia (20). Also, a recent case report showed that variants in SLCO1B3 (c.334T>G and c.699G>A) and ABCB11 (c.1331T>C) and the interaction between rosuvastatin and ticagrelor led to rhabdomyolysis in a patient with chronic kidney disease and other chronic conditions (21), but no other reports were found.
CYP3A5*3 may also have contributed to statin myotoxicity, since it has been associated with increased risk to atorvastatin and rosuvastatin-related myalgia in South-Indian dyslipidemic patients (22). However, this variant was not associated to statin intolerance in another study (23). Most studies have evaluated the effect of individual variants in SRAE, and not the interaction between a group of variants in key genes in statin pharmacokinetics pathway. Therefore, we suggest that the combined effect of the low-activity variants in SLCO1B1 and SLCO1B3, the high-activity variant in ABCB11, and the lack of activity of CYP3A5*3 predisposed the patient to low hepatic uptake, metabolization and efflux, respectively. The resulting higher rosuvastatin plasma concentration increased its systemic exposure, which may have caused myalgia (Figure 2).
Importantly, the patient carries LDLR rs28941776 (c.1646G>A, p.Gly549Asp), a disruptive-missense variant that showed reduced LDL uptake in an in vitro study (24). LDLR variants have been associated with variability in statin response in FH patients (25), but we did not find studies that investigated the association between LDLR variants and time to statin response or myalgia. Nevertheless, this variant could have played a role in patient’s rosuvastatin time to response and it should be considered for further studies.
A limitation of this study is that plasma concentrations of rosuvastatin and its metabolites were not measured. However, the adherence of the patient to the prescribed treatment was ensured using a validated adherence questionnaire and regular follow-up calls.
In summary, the combination of four low-activity variants in SLC genes, a high-activity variant in ABCB11, and a non-functional variant in CYP3A5 may explain the observed late response to rosuvastatin and the statin-related myalgia. With this case report, we have shown the importance of considering a combination of variants in a pharmacogenetic analysis to predict individual responses to statin treatment and prevent adverse drug events. We believe this study contributes to precision medicine in future clinical settings.
Patient perspective
“I have had high cholesterol since I was a child and it has been an issue because of the delayed response to treatments and of many adverse reactions to medications, especially simvastatin. The authors have been very attentive towards me throughout the whole study and discovered possible variants that may delay my response to rosuvastatin and influence the pain that I have felt when using statins. I am very happy for knowing the cause of my problem and I would like to thank the authors for this possible diagnosis. This has improved my perspectives of cholesterol treatment.”
Acknowledgments
The authors thank Adriana Garofalo, Dr. Hui Tzu Lin Wang, colleagues from the Laboratory of Molecular Investigation in Cardiology, and the Divisions of Dyslipidemia and Pharmacy of the Institute Dante Pazzanese of Cardiology. Their immeasurable technical and logistic support in patient selection and data collection made this study possible.
Funding: This work was supported by Sao Paulo Research Foundation (FAPESP), Brazil [Research grant: #2016/12899-6 to MHH]; and National Council for Scientific and Technological Development [CNPq, grant: #447120/2014-0 to MHH], Brazil. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. CDH is a recipient of a fellowship of the São Paulo Research Foundation (FAPESP), grant #2016/25637-0. RCCF, RHB, GMF and VFO are recipients of fellowships from FAPESP, Brazil. AAM is a recipient of fellowship from CAPES, Brazil. ESRM, MHH and RDCH are recipients of fellowships from CNPq, Brazil. BL was a recipient of fellowship from FAPESP, Brazil.
Footnote
Reporting Checklist: The authors have completed the CARE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-5540
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-5540
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-5540). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The intervention study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The DNA sequencing study was approved by the Ethics Committees of the Institute Dante Pazzanese of Cardiology (CAAE #4618713.0.1001.5462) and the School of Pharmaceutical Sciences of the University of Sao Paulo (CAAE #24618713.0.3001.0067), Sao Paulo, Brazil. The intervention study was approved by the Ethics Committee of the Institute Dante Pazzanese of Cardiology (CAAE #05234918.4.0000.5462). All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committees. The patient signed the written informed consents before her enrollment in the studies. In the written informed consent of the DNA sequencing study, the patient was informed that clinical data and blood samples would be collected for laboratory tests and genetic analyses. As for the intervention study, the patient was informed on the intervention protocol and sample collections throughout the visits, and that this data would be used for genetic and epigenetic analyses.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zhang Y, Cong H, Man C, et al. Risk factors for cardiovascular disease from a population-based screening study in Tianjin, China: a cohort study of 36,215 residents. Ann Transl Med 2020;8:444. [Crossref] [PubMed]
- Luvai A, Mbagaya W, Hall AS, et al. Rosuvastatin: A Review of the pharmacology and clinical effectiveness in cardiovascular Disease. Clin Med Insights Cardiol 2012;6:17-33. [Crossref] [PubMed]
- Rocha KC, Pereira BMV, Rodrigues AC. An update on efflux and uptake transporters as determinants of statin response. Expert Opin Drug Metab Toxicol 2018;14:613-24. [Crossref] [PubMed]
- Roden DM, McLeod HL, Relling MV, et al. Pharmacogenomics. Lancet 2019;394:521-32. [Crossref] [PubMed]
- Defesche JC, Lansberg PJ, Umans-Eckenhausen MA, et al. Advanced method for the identification of patients with inherited hypercholesterolemia. Semin Vasc Med 2004;4:59-65. [Crossref] [PubMed]
- Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405-24. [Crossref] [PubMed]
- Ben AJ, Neumann CR, Mengue SS. The Brief Medication Questionnaire and Morisky-Green test to evaluate medication adherence. Rev Saude Publica 2012;46:279-89. [Crossref] [PubMed]
- Stein EA, Strutt K, Southworth H, et al. Comparison of rosuvastatin versus atorvastatin in patients with heterozygous familial hypercholesterolemia. Am J Cardiol 2003;92:1287-93. [Crossref] [PubMed]
- Lee HK, Hu M, Lui SSh, et al. Effects of polymorphisms in ABCG2, SLCO1B1, SLC10A1 and CYP2C9/19 on plasma concentrations of rosuvastatin and lipid response in Chinese patients. Pharmacogenomics 2013;14:1283-94. [Crossref] [PubMed]
- Ho RH, Tirona RG, Leake BF, et al. Drug and bile acid transporters in rosuvastatin hepatic uptake: function, expression, and pharmacogenetics. Gastroenterology 2006;130:1793-806. [Crossref] [PubMed]
- Kameyama Y, Yamashita K, Kobayashi K, et al. Functional characterization of SLCO1B1 (OATP-C) variants, SLCO1B1*5, SLCO1B1*15 and SLCO1B1*15+C1007G, by using transient expression systems of HeLa and HEK293 cells. Pharmacogenet Genomics 2005;15:513-22. [Crossref] [PubMed]
- Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 2015;31:3555-7. [Crossref] [PubMed]
- Schwarz UI, Meyer zu Schwabedissen HE, Tirona RG, et al. Identification of novel functional organic anion-transporting polypeptide 1B3 polymorphisms and assessment of substrate specificity. Pharmacogenet Genomics 2011;21:103-14. [Crossref] [PubMed]
- Soko ND, Chimusa E, Masimirembwa C, et al. An African-specific profile of pharmacogene variants for rosuvastatin plasma variability: limited role for SLCO1B1 c.521T>C and ABCG2 c.421A>C. Pharmacogenomics J 2019;19:240-8. [Crossref] [PubMed]
- Hustert E, Haberl M, Burk O, et al. The genetic determinants of the CYP3A5 polymorphism. Pharmacogenetics 2001;11:773-9. [Crossref] [PubMed]
- Bailey KM, Romaine SP, Jackson BM, et al. Hepatic metabolism and transporter gene variants enhance response to rosuvastatin in patients with acute myocardial infarction: the GEOSTAT-1 Study. Circ Cardiovasc Genet 2010;3:276-85. [Crossref] [PubMed]
- Lee YS, Chun P. Effect of SLCO1B1 T521C on Statin-induced Myotoxicity: A Systematic Review and Meta-analysis. Korean J Clin Pharm 2018;28:320-30. [Crossref]
- Liu DJ, Peloso GM, Yu H, et al. Exome-wide association study of plasma lipids in >300,000 individuals. Nat Genet 2017;49:1758-66. [Crossref] [PubMed]
- Bai X, Zhang B, Wang P, et al. Effects of SLCO1B1 and GATM gene variants on rosuvastatin-induced myopathy are unrelated to high plasma exposure of rosuvastatin and its metabolites. Acta Pharmacol Sin 2019;40:492-9. [Crossref] [PubMed]
- Turner RM, Pirmohamed M. Statin-Related Myotoxicity: A Comprehensive Review of Pharmacokinetic, Pharmacogenomicand Muscle Components. J Clin Med 2019;9:22. [Crossref] [PubMed]
- Calderon-Ospina CA, Hernández-Sómerson M, García AM, et al. A pharmacogenomic dissection of a rosuvastatin-induced rhabdomyolysis case evokes the polygenic nature of adverse drug reactions. Pharmgenomics Pers Med 2020;13:59-70. [Crossref] [PubMed]
- Ramakumari N, Indumathi B, Katkam SK, et al. Impact of pharmacogenetics on statin-induced myopathy in South-Indian subjects. Indian Heart J 2018;70:S120-5. [Crossref] [PubMed]
- Fiegenbaum M, da Silveira FR, Van der Sand CR, et al. The role of common variants of ABCB1, CYP3A4, and CYP3A5 genes in lipid-lowering efficacy and safety of simvastatin treatment. Clin Pharmacol Ther 2005;78:551-8. [Crossref] [PubMed]
- Thormaehlen AS, Schuberth C, Won HH, et al. Systematic cell-based phenotyping of missense alleles empowers rare variant association studies: a case for LDLR and myocardial infarction. PLoS Genet 2015;11:e1004855. [Crossref] [PubMed]
- Choumerianou DM, Dedoussis GV. Familial hypercholesterolemia and response to statin therapy according to LDLR genetic background. Clin Chem Lab Med 2005;43:793-801. [Crossref] [PubMed]


