
N6-methyladenosine (m6A)-mediated messenger RNA signatures and the tumor immune microenvironment can predict the prognosis of hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) remains a high incidence malignancy with high mortality rate and poor prognosis worldwide. It has been reported that there are approximately 850,000 new HCC patients annually, with about 500,000 deaths due to HCC-associated diseases (1,2). Hepatitis B and C viral infections, excess alcohol consumption, aflatoxin B1 exposure, obesity, diabetes, and metabolic diseases are major risk factors for the development of HCC (3). Owing to the difficulty in making an early diagnosis, HCC patients are typically diagnosed at an advanced stage of the disease, and thus, treatments are generally associated with a low survival rate, high risk of recurrence, malignant metastasis, as well as a high risk of selection and spreading of drug resistance (4). Recently, immune therapies, such as immune checkpoint inhibition, have been used in advanced HCC to modify the carcinogenic process and to augment adaptive immunity (5). It has been shown that the tumor immune microenvironment (TIME), which is composed of both pro- and anti-tumor immune cells, can be reprogrammed by tumor-derived factors, which are involved in immune evasion and tumor progression (6,7). Accumulating evidence supports a role for the TIME in determining HCC progression, recurrence, metastasis, and poor outcome (8,9). Tumor-infiltrating immune cells (TIICs) are important components of the TIME and have been shown to be significant predictors of HCC survival (10). However, correlations between TIME parameters and HCC genesis and development are still poorly understood (10).
Currently, the most well studied messenger ribonucleic acid (mRNA) modification is N6-methyladenosine (m6A). It is the most pervasive internal mRNA modification (10), and correlates with tumor progression (11). Previous studies have shown that m6A demethylation of the fat mass and obesity-associated protein (FTO) is up-regulated and promotes tumor proliferation and metastasis in breast cancer (12,13). However, the role of the m6A modification in HCC tumorigenesis remains unclear.
We hypothesized that m6A may regulate HCC tumorigenesis and progression by regulating the immune system and thus may be a therapeutic drug target for HCC. To evaluate this hypothesis, data from The Cancer Genome Atlas (TCGA) were used as an effective cohort, and data from the Gene Expression Omnibus (GEO) database were used as a validation cohort. Genes associated with the immune microenvironment were downloaded from the Immunology Analysis Portal database (https//www.immport.org/shared/genelists), analyzed, and immune phenotypes were classified by immune scores. We validated the different m6A modification patterns and explored the relationship between immune infiltration and m6A regulation. We also identified three different immune phenotypes: the immune rejection phenotype, the immune inflammation phenotype, and the immune desert phenotype, and found that formation of the tumor microenvironment plays a role in the phenotype. We combined m6A modification patterns and immune infiltration phenotypes to establish a system to estimate HCC prognosis.
We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/atm-20-7396).
Methods
Data collection
The work-flow of this study is illustrated in Figure S1A. All related information was downloaded from TCGA data portal (http://cancergenome.nih.gov/). And the study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Data cohorts with missing information were removed. Data collection, refinement, and model statistics are summarized in Table S1. RNA sequence data from the National Cancer Institute’s (NCIs) Genomic Data Commons (GDC, https://gdc.cancer.gov/about-data/publications/pancanatlas), Fragments Per Kilobase per total Million mapped reads (FPKM), and TCGA biolinks were also used (14,15). Data were analyzed using the statistical software package R, version 3.6.1. Raw data were processed into R/Bioconductor.
Unsupervised gene cluster analysis
We used clustering, a resampling-based clustering algorithm, to analyze the m6A gene profiles of HCCs. A total of 21 m6A regulators, including eight writers, two erasers, and 11 readers were analyzed. We performed unsupervised cluster analysis with a consistent clustering algorithm and the Consensus Cluster Plus package as described previously (16).
Gene Set Variation Analysis (GSVA) and functional annotation
For an in-depth analysis of the biological processes that associate with the different m6A modification patterns, the “GSVA” R package for GSVA enrichment analysis was utilized. The gene set “c2.cp.kegg. v6.2. symbols” was downloaded from the MSigDB database and used to run GSVA analysis. For functional annotation of genes, we utilized cluster analysis and a false discovery rate <0.05.
Estimation of the immune cells infiltrating the TIME
A gene set of a variety of human immune cell subtypes was obtained from Charoentong et al. (17). Single-sample Gene Set Enrichment Analysis (ssGSEA) calculates separate enrichment scores for each sample and gene pair. This analysis showed the relative abundance of each cell type that infiltrated the TIME for each sample.
Classification of m6A phenotypes based on differential gene expression
To validate and classify m6A-associated genes, differentially expressed gene (DEG) analysis (fold-changes) and statistical computations of the two conditions were conducted using the EBSeq R package (https://bioconductor.org/packages/release/bioc/html/EBSeq.html).
Construction of m6A gene markers and m6A scoring
To validate m6A regulator modifications in HCC samples, we established a scoring system to estimate m6A gene characteristics. To establish m6A gene markers, DEGs of the different m6A clusters were normalized to extract overlapping genes, the unsupervised clustering method was used to analyze the overlapping data, and patients were divided into multiple groups for in-depth analysis. The number and stability of gene clusters were defined by the consistent clustering algorithm. We then used a univariate Cox regression model to analyze the association of each gene cluster with prognosis. We selected principal component 1 and principal component 2 as signature scores. Subsequently, we defined the m6a score using a method like that used in analyzing gene-gene interactions (GGIs): m6A Score= Σ (PC1i + PC2i).
Analysis of m6A modifications and their immune-related signatures in HCC
To investigate associations between m6A modifications and the immune microenvironment, we downloaded immune-related genes from the ImmPort database (http://www.immport.org), and identified prognostic gene signatures based on genes with independent prognostic values as determined previously (18). Our m6A scoring system revealed that m6A gene characteristics are associated with particular biological pathways.
Statistical analysis
DEGs were applied in this study with the threshold of absolute log2-fold-change >1 and an adjusted P value <0.05 using the R package “limma”. The overall survival rate was evaluated using the log-rank test with Kaplan-Meier estimation. Differences between the groups were evaluated by analysis of variance (ANOVA). All P values were two-sided, and P values <0.05 were considered significant. Comparative studies were analyzed using the Kruskal-Wallis test and Wilcoxon test. *P<0.05, **P<0.01, ***P<0.001. All statistics were calculated using R language.
Results
Expression of m6A regulators in HCC
To better understand the search strategy and selection of datasets for comprehensively understand m6A regulation in HCC progression, we constituted a flow chart of this study (Figure S1A). In total, 371 patients were involved in this study and three clusters were identified. The transcription levels of 21 m6A regulators, which included 11 readers, eight writers, and two erasers, as well as the copy number variations (CNVs) and somatic mutations of these regulators, were described in a previous study (19). The chromosomal locations of the amplification mutations were distributed randomly and unequally across the chromosomes (Figure 1A). Of 64 HCC samples, 44 contained mutations related to m6A modifications (68.75% mutation rate). The most prevalent mutation were found in the KIAA1429 (8%) and LRPPRC (8%) genes, followed by HNRNPC (6%) and YTHDC2 (6%) (Figure 1B). The 21 m6A regulators exhibited widespread gene gain and loss mutations, KIAA1429 and YTHDF3 contained the most gain mutation type, while ALKBH5 and ZC3H13 had the most frequent loss mutation type (Figure 1C). We also explored the 21 regulators in tumor tissue compared with normal tissue, and easily identified the two subgroups; the red spot represented normal tissue and the blue spot represented tumor tissue (Figure 1D).

To further investigate the overall survival between the regulators, HCC patients were divided into mutated subgroup and wide type subgroup based on whether they were combined with m6A mutation information. Obviously, the mutated subgroups had a poor prognosis in YTHDF1 [P=0.018, hazard ratio (HR) =4.7], LRPPRC (P=0.003, HR =6.44), and FTO (P=4.12e7, HR =11.3) (Figure S1B). We also performed a meta-analysis, and the results revealed that the regulators have prognostic potential in HCC cohorts (Figure S1C). The mean expression levels of m6A regulators in HCC specimens and normal tissues are shown in Figure 1E; the difference was significant and m6A regulators were highly expressed in both tumor tissue and normal tissue. In this study, we mapped the gene somatic mutation information of 21 m6A regulators between tumor tissue and normal tissue, and confirmed that m6A modification plays an important role in HCC tumorigenesis and progression.
m6A regulators have close relationship with cellular crucial functional pathways regulation
In order to comprehensively explain the crosslink among the 21 m6A regulators, we divided 21 m6A regulators into three clusters as illustrated in Figure 2A. Through GSVA of functional genes, we determined the biological roles of the m6A regulators. The expression of genetic information of the m6A regulators was controlled through intricate regulatory networks and a complex regulatory relationship. The overall survival curve implied a significant survival advantage for cluster 2 (*P<0.0001, Figure 2B).
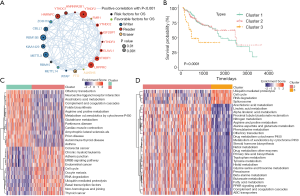
In the present study, we explored the relationship between different modification patterns and biological functional activities. The results indicated that three clusters exhibit significant pathway enrichment. The cluster 1-associated pathways gather on translation functions, such as olfactory transduction, neuroactive ligand receptor interaction, arachidonic acid metabolism complements and coagulation cascades, folate biosynthesis, arginine and proline metabolism, metabolism of xenobiotics by cytochrome, glutathione metabolism and Parkinson’s disease cardiac muscle contraction, amyotrophic lateral sclerosis, prion diseases, and autoimmune diseases. Cluster 2 functional pathways were significantly enriched in the ERBB signaling pathway, cell cycle signaling pathway, cell adhesion, as well as other pathways related to cell matrix adhesion and carcinogenesis, such as colorectal cancer, chronic myeloid leukemia, adherent’s junction, the ERBB signaling pathway, endometrial cancer, the cell cycle, oocyte meiosis, RNA degeneration, ubiquitin mediated proteolysis, basal transcription factors, non-homologous end joining, and thyroid cancer (Figure 2C). The third cluster-associated functional pathways act in metabolism pathways (Figure 2D). Our results implied that m6A regulators participated in many important cellular activities, including cellular metabolism regulation, nuclear factor regulation, signal transitional regulation, and carcinogenesis.
m6A regulators have a close relationship with the infiltration of immune cells
Next, we estimated the enrichment score of immune cells among the three clusters and identified the differences in the infiltration of immune cells among clusters 1–3. The box plot exhibited significant differences in most immune cells, including activated B cells, activated cluster of differentiation (CD)4+T cells, activated CD8+T cells, ᵞᵟT cells, regulatory T cells, type 1 helper cells, type 17 helper cells, activated dendritic cells (DCs), CD56 bright natural killer (NK) cells, CD56 dim NK cells, eosinophils, macrophages, and mast cells (Figure 3A).
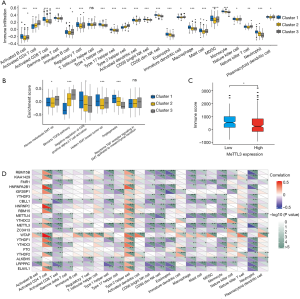
To further investigate the biological behaviors among the three m6A modification clusters, pathway enrichment analysis was applied. Our results indicated that the transforming growth factor beta (TGF-β) pathway, angiogenesis, and the tumor EMT process were remarkably different among the clusters (Figure 3B). Our research suggested that the three types of regulators were involved in crucial functional regulations with different pathways. According to the GSVA analysis, the 21 m6A regulators were classified as either an immune rejected phenotype, an immune inflamed phenotype, or an immune desert phenotype based on the infiltration of immune cells (Figure 3A,B).
We then focused on methyltransferase like 3 (METTL3), and the immune score of different METTL3 expression subgroups showed a weak difference (Figure 3C). Furthermore, Spearman correlation analysis was performed for TIME infiltration and the m6A regulators (Figure 3D). METTL3 expression correlated with many of the TIME infiltrating immune cells and the immune score. Our study implied that the infiltration of CD4+ T cells, type 1 T helper cells, type 2 T helpers, CD56 bright NK cells, eosinophils, macrophages, mast cells, monocytes, and neutrophils exhibited a significant difference between high and low METTL3 expression (Figure S2A), indicating that low METTL3 expression resulted in increased TIME-associated immune cell infiltration.
Subsequently, we estimated the gene expression of immune checkpoint molecules and receptors, and discovered that CD80, CD86, intercellular adhesion molecule (ICAM) 1, ICAM3, and programmed cell death ligand 1 (PDL1) gene expression showed differences across different METTL3 expression levels (Figure S2B). We then evaluated the enrichment score of some crucial cell activity pathways between two clusters, and the box plot exhibited significant differences in the TGF-β pathway, T cell activation, angiogenesis, and the tumor EMT process (Figure S2C). These results indicated that the m6A regulators play key roles in immune cell infiltration and characteristic TIME formation.
The transcriptome and clinical features of m6A regulators
To further identify the cellular biological behavior and in-depth mechanisms of the m6A regulators, we performed unsupervised cluster analysis on m6A phenotype-related genes to divide patients into one of four different clusters based on the m6A modified genomic phenotypes (Figure 4A). Based on the m6A regulator classifications, we confirmed that cluster 1, with the abundant immune cell infiltration subgroup, exhibited a better prognosis and survival probability (Figure 4B). Among the three immune cell infiltration-associated subgroups, expression of regulatory factors of 21 regulators differed significantly (Figure 4C).
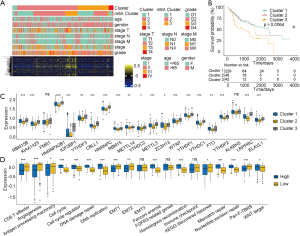
Further analysis of the matrix-related pathways revealed that the gene expression level of the m6A regulators suggested significantly different expression levels of immune related functional annotation and activation of matrix pathways, including CD8+T cells effector, angiogenesis, the cell cycle, cell cycle regulator, deoxyribonucleic acid (DNA) damage repair, DNA replication, EMT1, Fanconi anemia, homologous recombination, mismatch repair, nucleotide excision repair, and Pan-F-TBRS (Figure 4D). Here, we confirmed that the 21 regulators were classified into three gene clusters based on gene expression and cluster analysis. The box plots illustrated that the 21 regulators gene expression clusters were consistent with the functional classification.
The m6A regulators gene expression classification with immune infiltration
To better annotate the functions of m6A-related genes with immune cells, our analysis results implied that the infiltration of activated CD4+ T cells, regulatory T cells, type 17 T helper cells, CD56 bright NK cells, CD56 dim NK cells, and eosinophils exhibited significant differences between the three clusters (Figure 5A). To explore the roles of m6A-related phenotypes in the modulation of the TIME, we analyzed the expression of chemokines and cytokines in the three m6A gene clusters. Interferon gamma (IFN-γ), CD8A, T-Box transcription factor 2 (TBX2), and tumor necrosis factor (TNF) showed significant differences among the three clusters. Other cytokines exhibited no or weak differences between these clusters (Figure 5B). In Figure 5C,D, we estimated the gene expression of immune checkpoint molecules and receptors and found that HAVCR2, CD80, PDCD1, TIGIT, CD86, COL4A1, ZEB, TGFB2, and TWIST1 gene expression showed remarkable differences between the three clusters, which suggested that these immune checkpoints had a significant relationship with the m6A regulator classification.
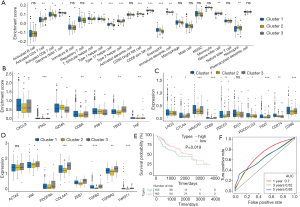
We further investigated the prognostic value of the m6A scores for HCC. HCC patients were divided into high and low m6A score groups, and the survival of these groups was compared. The overall survival for the low score m6A group was better than the high score group [P=0.019, HR 1.6 (1.1–2.3)]. The 5-year survival rate was 56% for the high m6A score group and 48% for the low score group (Figure 5E). The area under the curve (AUC) in patients treated with immunotherapy was 0.768 (Figure 5F). Our data demonstrated that the m6A evaluation scores based on modification patterns were correlated with immune or matrix activation, which affects tumor immune infiltrates and patient prognosis.
Discussion
Globally, HCC is still a lethal malignancy with a poor prognosis. The tumorigenesis mechanism of HCC must be elucidated to improve prognosis, identify biomarkers, and develop effective therapeutic strategies. The liver is the largest immune-related organ, and the immune system plays a definitive role in oncogenesis (20,21). Recent studies have indicated that aberrant mRNA modifications contribute to HCC prognosis and overall survival. Currently, m6A is the most well studied post-transcriptional mRNA modification, and evidence suggests that this modification epigenetically affects mRNA processing, translation, and stability (22). Dysregulation of the m6A modification has been shown to be associated with the initiation and progression of various cancers. Thus, exploring the relationship between m6A regulation and the TIME could lead to the design of immune-based targeted therapies for HCC.
METTL3 is a crucial oncogene in several cancers and has been shown to be up-regulated in gastric cancer (23), prostate carcinoma (24), and osteosarcoma (25). Also, methyltransferase like 14 (METTL14) has been shown to play a crucial role in the RNA epigenetics of colorectal cancer, and thus, has prognostic value (26). It is also been shown that YTHDF2 suppresses cancer cell migration via m6A-associated modification (26). In the present study, we identified three different m6A modification patterns and their relationship with the infiltration of immune cells, such as NK cells, macrophages, eosinophils, mast cells, myeloid-derived suppressor cells (MDSCs), and plasma cell-like DCs, into the HCC microenvironment. Based on the infiltration of immune cells and the immune score, we identified three different immune-associated phenotypes of the HCC tumor environment: immune excluded, immune inflamed, and immune desert. The immune excluded phenotype is characterized by the presence of T cells at the periphery of cancer nests. In the immune excluded phenotype of bladder cancer, there was no PD-L1 expression and only a few tumor-infiltrating lymphocytes (TILs) were present (27). Mariathasan et al. found that combination therapy with TGF-blocking and anti-PD-L1 antibodies decreased TGF signaling in stromal cells, facilitated concentration of T cells in the centers of tumors, and provoked vigorous anti-tumor immunity and tumor regression (28). Zhao et al. demonstrated that the immune inflamed phenotype of triple-negative breast cancers is characterized by the infiltration of CD8+ T cells into the tumor parenchyma, indicating that these patients are more likely to benefit from immune checkpoint inhibitor therapy (29). Job et al. reported that the inflamed subtype is characterized by massive T lymphocyte infiltration as well as activation and upregulation of inflammatory and immune checkpoint pathways. This phenotype is associated with a prolonged survival prognosis (30). Thus, patients with the immune inflamed phenotype may benefit from immunotherapy. In this study, we found that the immune desert phenotype is characterized by CD8 cell deficiency and is associated with a poor prognosis.
The TIME can predict patient survival in most cancer types, and m6A modification has been shown to affect cell infiltration in the TIME and the epigenetic regulation of the immune response in gastric cancer. M6A has also been shown to regulate the innate immune response by targeting type I interferons (18,31). In the innate immune system, neoantigen-specific immunity is regulated by mRNA methylation via the m6A-binding protein YTHDF1 (32). In this study, we explored m6A regulatory factor-mediated TIME cell infiltration and found more DCs, including activated DCs, immature DCs, and plasma cell-like DCs in tumors with low METTL3 expression, indicating that METTL3-mediated m6A methylation modification activates DCs in the TIME, thereby enhancing the anti-tumor immune response.
Conclusions
Our findings demonstrated that numerous immune-related genes regulated by m6A modification contribute to the immune phenotype. Moreover, we validated a novel model of m6A modification and immune-related gene regulation to predict HCC prognosis. The immune score and immune classification systems may lead to novel therapeutic targets for HCC.
Acknowledgments
We thank the patients and investigators who participated in TCGA and the GEO for providing the data analyzed in this study.
Funding: This study was supported by funds from the National Natural Science Foundation of China (No 81902832); the Science and Technology Research Project of Henan Province (No 192102310117 and 202102310115); and the Gandan Xiangzhao Research Fund (No GDXZ2019001 and GDXZ2019007).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-7396
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-7396). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kempinska K, Malik B, Borkin D, et al. Pharmacologic Inhibition of the Menin-MLL Interaction Leads to Transcriptional Repression of PEG10 and Blocks Hepatocellular Carcinoma. Mol Cancer Ther 2018;17:26-38. [Crossref] [PubMed]
- Llovet JM, Zucman-Rossi J, Pikarsky E, et al. Hepatocellular carcinoma. Nat Rev Dis Primers 2016;2:16018. [Crossref] [PubMed]
- Yang JD, Hainaut P, Gores GJ, et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol 2019;16:589-604. [Crossref] [PubMed]
- Xie QL, Liu Y, Zhu Y. Chromosome region maintenance 1 expression and its association with clinical pathological features in primary carcinoma of the liver. Exp Ther Med 2016;12:59-68. [Crossref] [PubMed]
- Ou Q, Yu Y, Li A, et al. Association of survival and genomic mutation signature with immunotherapy in patients with hepatocellular carcinoma. Ann Transl Med. 2020;8:230. [Crossref] [PubMed]
- Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol 2013;14:1014-22. [Crossref] [PubMed]
- Azambuja JH, Ludwig N, Braganhol E, et al. Inhibition of the Adenosinergic Pathway in Cancer Rejuvenates Innate and Adaptive Immunity. Int J Mol Sci 2019;20:5698. [Crossref] [PubMed]
- Carone C, Olivani A, Dalla Valle R, et al. Immune Gene Expression Profile in Hepatocellular Carcinoma and Surrounding Tissue Predicts Time to Tumor Recurrence. Liver Cancer 2018;7:277-94. [Crossref] [PubMed]
- Zhang FP, Huang YP, Luo WX, et al. Construction of a risk score prognosis model based on hepatocellular carcinoma microenvironment. World J Gastroenterol 2020;26:134-53. [PubMed]
- Hsiao YW, Chiu LT, Chen CH, et al. Tumor-Infiltrating Leukocyte Composition and Prognostic Power in Hepatitis B- and Hepatitis C-Related Hepatocellular Carcinomas. Genes (Basel) 2019;10:630. [Crossref] [PubMed]
- Zhao X, Chen Y, Mao Q, et al. Overexpression of YTHDF1 is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Biomark 2018;21:859-68. [Crossref] [PubMed]
- Han M, Yang G, Lin Q, et al. Determination of Endogenous Bufalin in Serum of Patients With Hepatocellular Carcinoma Based on HPLC-MS/MS. Front Oncol 2020;9:1572. [Crossref] [PubMed]
- Yang Z, Li J, Feng G, et al. MicroRNA-145 Modulates N6-Methyladenosine Levels by Targeting the 3'-Untranslated mRNA Region of the N6-Methyladenosine Binding YTH Domain Family 2 Protein. J Biol Chem 2017;292:3614-23. [Crossref] [PubMed]
- Colaprico A, Silva TC, Olsen C, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res 2016;44:e71. [Crossref] [PubMed]
- Grossman RL. Data Lakes, Clouds, and Commons: A Review of Platforms for Analyzing and Sharing Genomic Data. Trends Genet 2019;35:223-34. [Crossref] [PubMed]
- Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 2010;26:1572-3. [Crossref] [PubMed]
- Charoentong P, Finotello F, Angelova M, et al. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep 2017;18:248-62. [Crossref] [PubMed]
- Zhang B, Wu Q, Li B, et al. m6A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol Cancer 2020;19:53. [Crossref] [PubMed]
- Qin L, Min S, Shu L, et al. Genetic analysis of N6-methyladenosine modification genes in Parkinson's disease. Neurobiol Aging 2020;93:143.e9-143.e13. [Crossref] [PubMed]
- Angell H, Galon J. From the immune contexture to the Immunoscore: the role of prognostic and predictive immune markers in cancer. Curr Opin Immunol 2013;25:261-7. [Crossref] [PubMed]
- Tang X, Shu Z, Zhang W, et al. Clinical significance of the immune cell landscape in hepatocellular carcinoma patients with different degrees of fibrosis. Ann Transl Med 2019;7:528. [Crossref] [PubMed]
- Zhao BS, Wang X, Beadell AV, et al. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 2017;542:475-8. [Crossref] [PubMed]
- Guan K, Liu X, Li J, et al. Expression Status And Prognostic Value Of M6A-associated Genes in Gastric Cancer. J Cancer 2020;11:3027-40. [Crossref] [PubMed]
- Yuan Y, Du Y, Wang L, Liu X. The M6A methyltransferase METTL3 promotes the development and progression of prostate carcinoma via mediating MYC methylation. J Cancer 2020;11:3588-95. [Crossref] [PubMed]
- Ling Z, Chen L, Zhao J. m6A-dependent up-regulation of DRG1 by METTL3 and ELAVL1 promotes growth, migration, and colony formation in osteosarcoma. Biosci Rep 2020;40:BSR20200282. [Crossref] [PubMed]
- Yang X, Zhang S, He C, et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer 2020;19:46. [Crossref] [PubMed]
- Mandelkow T, Blessin NC, Lueerss E, et al. Immune Exclusion Is Frequent in Small-Cell Carcinoma of the Bladder. Dis Markers 2019;2019:2532518. [Crossref] [PubMed]
- Mariathasan S, Turley SJ, Nickles D, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018;554:544-8. [Crossref] [PubMed]
- Zhao S, Ma D, Xiao Y, et al. Molecular Subtyping of Triple-Negative Breast Cancers by Immunohistochemistry: Molecular Basis and Clinical Relevance. Oncologist 2020;25:e1481-91. [Crossref] [PubMed]
- Job S, Rapoud D, Dos Santos A, et al. Identification of four immune subtypes characterized by distinct composition and functions of tumor microenvironment in intrahepatic cholangiocarcinoma. Hepatology 2019;72:965-81. [Crossref] [PubMed]
- Winkler R, Gillis E, Lasman L, et al. m6A modification controls the innate immune response to infection by targeting type I interferons. Nat Immunol 2019;20:173-82. [Crossref] [PubMed]
- Han D, Liu J, Chen C, et al. Anti-tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells. Nature 2019;566:270-4. [Crossref] [PubMed]


