
Distinctive genomic characteristics in POLE/POLD1-mutant cancers can potentially predict beneficial clinical outcomes in patients who receive immune checkpoint inhibitor
Introduction
The recent clinical success of immune checkpoint inhibitors (ICIs) has paved an entirely new avenue for cancer treatment. To date, numerous ICI agents have been approved to treat a variety of tumors (1). Despite ICIs representing a significant breakthrough in cancer therapy, there are still several challenges that limit their application (2-5), including the failure of most recipients to respond to treatment. Biomarkers, such as the expression of programmed death-ligand 1 (PD-L1), high microsatellite instability (MSI-H), and tumor mutational burden (TMB) have shown promising value for predicting the prognosis of ICI-treated patients and have been explored extensively (6-9). However, discrepancies exist between different studies.
High fidelity of DNA replication in eukaryotes, the prerequisite for preventing mutagenesis and tumor formation, is attributable to the combination of highly accurate DNA replication and exonuclease proofreading by the DNA polymerases Polδ and Polε, and post-replication correction by the DNA mismatch repair (MMR) system (10). The POLE and POLD1 genes encode the major catalytic and proofreading subunits of the Polε and Polδ enzyme complexes, respectively. DNA-repair deficiencies resulting from germline or somatic mutations in the POLE/POLD1 exonuclease (A.K.A. proofreading) domain have been shown to contribute to remarkably ultramutated phenotypes in endometrial and colorectal cancers (11-14). The most common POLE mutations identified in tumor genome include P286R/H, V411L and S459F; whereas very few somatic POLD1 mutations have been found (15,16). It has been reported that 1–3% of colorectal cancers and approximately 7% of endometrial cancers harbored POLE mutation (12,17). Preliminary clinical studies have also revealed the association between POLE/POLD1 proofreading mutations and beneficial clinical outcomes to ICI therapy in endometrial cancer (18), non-small cell lung cancer (19), colorectal cancer (20), and cervical carcinosarcoma (21). A recent study investigated the prevalence of POLE/POLD1 mutations across multiple cancer types in of 47,721 patients from cBioPortal database and demonstrated a potential predictive value of POLE/POLD1 mutations for positive outcomes to ICI treatment (22). However, functional evidences have been reported for only a small number of hotspot mutations in or close to the proofreading domains of POLE and POLD1; thus, the effects of the non-hotspot mutations on TMB and clinical outcomes remain elusive (23).
Tumors with deficient MMR function often present with a high frequency of insertion/deletion in short repetitive sequences in the genome, which is known as microsatellite instability (MSI) event. MSI occurs in approximately 15% of sporadic colorectal cancers (24), and also has been observed in endometrial, ovarian, stomach and urinary tract cancers (25). POLE/POLD1-mutated cancers are primarily microsatellite stable (MSS) (26,27), probably because cells cannot sustain an excessive accumulation of mutations caused by the simultaneous loss of proofreading and MMR functions (28,29). However, recent studies revealed that a subset of POLE-mutated endometrial and colorectal cancers also display MSI-H (30,31). How POLE/POLD1 proofreading and MMR deficiencies interact and drive hypermutation in such cancers has not been well-elucidated and merits further exploration.
In the present study, we retrieved the genomic data of 21,074 patients with various cancers from an in-house database, with the aim of investigating the prevalence and spectra of POLE and POLD1 mutations, and their correlation with TMB and MSI in Chinese population. We also explored their prognostic value for ICI treatment and association with tumor immune infiltration by extracting and re-analyzing data from published studies.
We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/atm-20-7553).
Methods
Study design, patients, and samples
The study design is detailed in Figure S1. The genomic data of 21,074 Chinese patients with 23 types of cancers were retrieved from the Burning Rock variant database (BR VarDB). Cancers included lung cancer (66.9%), colorectal cancer (11.0%), breast cancer (5.3%), gastric cancer (2.7%), ovarian cancer (1.5%), sarcoma (1.1%), liver cancer (0.9%), pancreatic cancer (0.8%), cervical cancer (0.7%), esophageal cancer (0.7%), head-neck cancer (0.7%), kidney cancer (0.6%), endometrial cancer (0.5%), biliary duct cancer (0.5%), melanoma (0.3%), gallbladder cancer (0.3%), prostate cancer (0.3%), GIST (0.3%), bladder cancer (0.2%), glioma (0.2%), thyroid cancer (0.1%), lymphoma (0.04%), and ureteral cancer (0.02%) (Figure S2A).
Samples were derived from participating hospitals and had been sequenced with one of the following panels (Burning Rock, Guangzhou, China): OncoScreen Plus (520 cancer-related genes and MSI loci), ColonCore (36 colorectal cancer-related genes and MSI loci), or LungPlasma (168 lung cancer-related genes) in a Clinical Laboratory Improvement Amendments (CLIA)/CAP-certified laboratory. The three panels have identical coverage for POLD1/POLE. Plasma samples with a maximum allele frequency (max AF) <0.5% were excluded. A total of 21,074 samples (1 from each patient), including 11,380 tumor tissue, 8,640 plasma, 843 pleural fluid, and 211 cerebrospinal fluid samples, were retained for further analysis. Detailed characteristics of the BR VarDB samples are shown in Figure S2. Additionally, POLE and POLD1 mutational data from 10,967 samples spanning 16 cancer types were obtained from The Cancer Genome Atlas (TCGA) database for comparison (https://www.cancer.gov/tcga). TMB was evaluated for 11,801 BR VarDB samples sequenced with the OncoScreen Plus panel, and MSI status was determined for 13,487 samples sequenced by the OncoScreen Plus or ColonCore panel. Subsequently, the associations of POLE/POLD1 mutation with TMB and MSI status were assessed.
The mutational profiles and clinical outcomes of 2,911 patients who received treatment with ICIs were retrieved from published data (1,910 pan-cancers (32,33), 429 NSCLCs (NCT01903993& (NCT02008227) (34), and 148 melanomas (35,36). The associations between POLE/POLD1 mutations and clinical outcomes including overall survival (OS) and overall response rate (ORR) were analyzed.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the institutional review board (IRB) of The First Affiliated Hospital of Zhengzhou University. Informed consent was not required due to the retrospective nature of the study.
Variant calling and mutational signature characterization
FASTQ format data were mapped to the reference human genome (hg19) using the Burrows-Wheeler Aligner v.0.7.10 (37) with default parameters. The Genome Analysis Tool Kit v.3.2 (38) and VarScan v.2.4.3 (39) were used for local alignment optimization, duplication marking, and variant calling. Base calling required at least 8 supporting reads for single nucleotide variations and 2 and 5 supporting reads for insertion-deletion variations, respectively. Variants with a depth <100 or population frequency >0.1% in the databases (ExAC, 1,000 Genomes, dbSNP, or ESP6500SI-V2) were filtered out and excluded from further analysis. The remaining variants were annotated with ANNOVAR (2016-02-01 release) (40) and SnpEff v.3.6 (41). DNA translocation analysis was performed using Factera v.1.4.3 (42). The copy number variation (CNV) was called with an in-house algorithm based on sequencing depth. COSMIC-reported mutational signatures in the tumor genome were quantified using the MuSiCa tool (http://bioinfo.ciberehd.org/GPtoCRC/en/tools.html) (43).
Assessment of TMB and MSI
TMB was computed for each patient as the ratio between the total number of nonsynonymous mutations detected with the total size of the coding region of the panel using the formula below. Mutations occurring on the kinase domain of epidermal growth factor receptor (EGFR) or anaplastic lymphoma kinase (ALK) were excluded from the mutation count.
The MSI status was determined based on a read-count distribution approach previously described (44). Briefly, each microsatellite locus was characterized with the coverage ratio of a specific set of repeat lengths and categorized as unstable if the coverage ratio was less than [mean − 3 × SD] of the reference ratio. A tumor sample was determined as MSI-H if more than 40% of the marker loci were length-instable, MSS if the percentage of length-instable loci were <15%, or MSI-L for if the percentage was between 15% and 40%.
Statistical analysis
Statistical analysis was performed using R version 3.3.3 software (Lucent Technologies, New Jersey, USA). Differences between POLE/POLD1 mutant and wildtype groups were calculated and presented using Fisher’s exact test, paired two-tailed Student’s t-test, or analysis of variance, as appropriate. Kaplan-Meier analysis was performed to estimate survival functions, and differences in the survival curves between groups were determined using the log-rank test. Cox multivariate proportional-hazards analysis was carried out to investigated potential predictors of survival. A P value of P<0.05 was considered to be statistically significant.
Results
The prevalence and spectra of POLE and POLD1 mutations
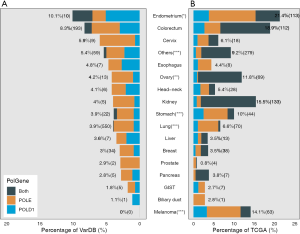
First, the prevalence of POLE and POLD1 mutations was compared between samples from BR VarDB and the TCGA database. All 16 TCGA cancer types were included in the BR VarDB. POLE/POLD1 mutation(s) were detected in 4.4% of the BR VarDB samples compared to 4.35% of samples from the TCGA database (P=0.845). POLE was mutated in 3.2% of tumors in both the BR VarDB and TCGA datasets, while POLD1 mutation was present in 1.4% and 1.6% of BR VarDB and TCGA tumors, respectively (P=0.248). POLE and POLD1 co-mutation was slightly more common among TCGA tumors than in BR VarDB tumors (0.43% vs. 0.21%, P<0.001). In BR VarDB samples, POLE / POLD1 mutations were most commonly seen in cancers of the endometrium (10.1%), colorectum (8.3%), cervix (5.9%), esophagus (4.8%), and ovary (4.2%) (Figure 1), whereas POLE/POLD1 mutations exhibited the highest frequency in endometrial cancer (19.1%), melanoma (12.3%), stomach adenocarcinoma (9.1%), colorectal cancer (8.8%), and lung cancer (6.4%) in the TCGA dataset. Compared with TCGA, the BR VarDB dataset showed significantly lower rates of POLE/POLD1 mutations in endometrial cancer (P=0.031), stomach adenocarcinoma (P<0.001), lung cancer (P<0.001), and melanoma (P<0.001) but a higher rate of the mutations in ovarian cancer (P<0.01).
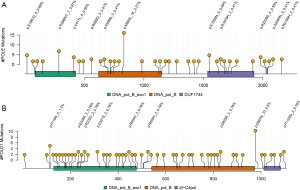
Next, we explored the spectra of POLE and POLD1 mutations identified in the BR VarDB. Of the 670 POLE mutations 78.5% were missense, and 11%, 5.2%, 4.3%, and 0.9% were splicing, frameshift, stop-gained variants, and in-frame indel mutations, respectively. Of the 331 POLD1 mutations 81.9%, 5.7%, 8.8%, and 3.0% were missense, splicing, frameshift, and stop-gained mutations, respectively. Of the 928 POLE/POLD1 mutation types in total, 130 and 798 were in the proofreading and non-proofreading domain, respectively. The mutation rates of POLE (proof vs. non-proof: 0.28 vs. 0.32 mut/aa, P=0.82) and POLD1 (proof vs. non-proof: 0.34 vs. 0.39 mut/aa, P=0.79) were comparable in the proofreading domain versus the non-proofreading domain. p.M820L was the most commonly occurring mutation in POLE across all cancer types (n=16), as well as in lung cancer (n=13), followed by P286R/T (n=7), E18K/Q (n=5), and K1550fs (n=5) (Figure 2A). p. D987fs/N (n=10) was the hotspot mutation observed in POLD1 (Figure 2B).
The distinctive mutational signatures in POLE/POLD1-mutated tumors
To obtain a deeper insight into the distinct mechanism of mutations in the genome of POLE/POLD1-mutated tumors, we compared the mutational signatures of POLE/POLD1-mutated and POLE/POLD1 wild-type (WT) tumors. First, we incorporated the sequence context in which substitution mutation occurred, by considering the bases 5’-and 3’-flanking to each mutated base (Figure 3A). Six types of base substitution and 16 possible sequence contexts for each mutated base generated 96 possible mutated trinucleotides. The two groups displayed similar patterns of contexts with some subtle differences. POLE/POLD1-mutated tumors harbored more TCT→TAT and TCG→TTG transversions than POLE/POLD1 WT tumors, which manifested as a distinctive missense and truncation mutational pattern in oncoproteins and tumor suppressors (27). POLE/POLD1-mutated tumors also had fewer CTG→CGG and TCA→TTA transversions than POLE/POLD1 WT tumors.

Next, we interrogated COSMIC-reported mutational signatures that had been well deciphered to correlate with specific mutational processes (Figure 3B). The signatures of tumors with POLE/POLD1-mutations were more predominantly associated with DNA MMR deficiency and ultra-hypermuation than those of POLE/POLD1 WT tumors. In contrast, the mutational signature of homologous recombination deficiency was only enriched in POLE/POLD1 WT tumors, which also exhibited elevation of signatures associated with age and smoking.
POLE/POLD1 mutation affected TMB and MSI status in tumors
Next, we investigated the correlation of POLE/POLD1 mutation with TMB. Tumors were classified into three groups based on the presence and location of the POLE/POLD1 mutation (Figure 4A). Tumors with a proofreading mutation in POLE/POLD1 had a significantly higher TMB than tumors with non-proofreading mutations (P<0.01), and both possessed a higher TMB than POLE/POLD1 WT tumors (P<0.0001 and P<0.0001, respectively). The same tendency was observed in the subsets of POLE-mutated tumors and POLD1-mutated tumors, which suggested that mutations occurring in the proofreading domains of the two genes affect the TMB more than those in other locations.
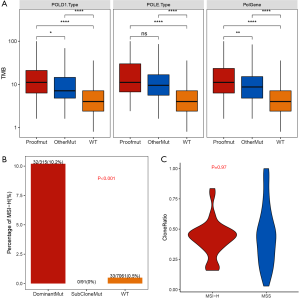
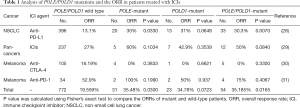
To identify specific POLE/POLD1 mutations that might serve as drivers for hypermutation, mutations occurring only in hypermutated (defined as TMB >50/Mb) and MSS tumors were investigated (Table 1). A total of 14 POLE and 9 POLD1 mutations were identified in 16 tumors. Among them, POLE p.V411L (n=2), p.P286R (n=1), p.Q292* (n=1), and p.D406N (n=1) were located in the proofreading domain, and tumors harboring these mutations had an extremely high TMB (>100/Mb). Except for p.D406N (AF =0.77%), all of these proofreading mutations appeared with high abundances in plasma (AF range, 7.8–16%). No POLD1 proofreading mutations were identified in hypermutated tumors. Two of the 9 POLD1 mutations, p.P269S (AF: 4.6%) and p.E245K (AF: 1.18%), were located close to the upstream of the proofreading domain.
Full table
To elucidate the interaction and effects of POLE/POLD1 mutation and MSI in tumorgenesis, tumors were stratified into different categories according to the clone-ratio of POLE/POLD1 mutation, which was defined as the mutational abundance normalized by maximum allelic fraction (AF) in the specific sample (AF/max AF). If the POLE/POLD1 mutation was a driver of the hypermutation and occurred before MSI, then tumors would accumulate the dominant clone of the driver mutation; conversely, if the POLE/POLD1 mutation appeared as passenger of MSI, then it would occur at a much lower ratio. Interestingly, tumors harboring dominant POLE/POLD1 mutation (AF/maxAF> 0.1) were remarkably more likely to be MSI-H than POLE/POLD1 WT tumors (10.2% vs. 0.5%, P<0.001). Moreover, all the tumors harboring sub-cloned POLE/POLD1 mutation (AF/max AF ≤0.1) were determined to be MSS (Figure 4B). The clone-ratio of POLE/POLD1 mutation was comparable between MSI-H and MSS tumors (P=0.97, Figure 4C).
The prognostic value of POLE/POLD1 mutation in ICI-treated cancer patients
Published mutational data and clinical survival information from 1661 ICI-treated patients with various cancers were retrieved (32), and the prognostic value of POLE/POLD1 mutation was subsequently investigated. Univariate analysis revealed POLE/POLD1-mutant patients to have a longermedian OS vs. POLE/POLD1-WT patients (34 vs. 17 months, P=0.0016, Figure 5A). However, the positive prognostic role of POLE/POLD1 mutation was not found to be significant in the multivariate analysis (HR =0.86, 95% CI: 0.62–1.19, P=0.372, Figure 5B).
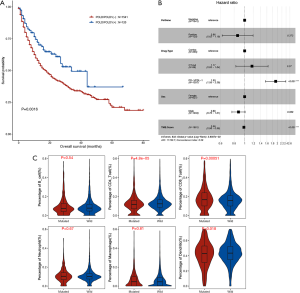
The ORR for ICI therapy was calculated and compared between POLE/POLD1-mutant and WT patients from four studies that provided information on both mutations and best tumor response (Table 1). Two of the studies reported a trend of a higher ORR in patients with POLE/POLD1 mutations. Analysis of the pooled data revealed a significantly higher ORR in POLE/POLD1-mutant patients than in POLE/ POLD1 WT patients (35.2% vs. 19.6%, P=0.0165).
Moreover, we investigated the association of POLE/POLD1 mutation with tumor microenvironment using the data derived from TCGA database (N=10,468). A significantly elevated fraction of CD8+ tumor-infiltrating lymphocytes (TILs) (17.1% vs. 15.9%, P=5.1×10−4) and a lower fraction of CD4+ TILs (11.8% vs. 12.4%, P=4.8×10−5) were observed in POLE/POLD1-mutant tumors compared to POLE/POLD1 WT tumors (Figure 5C).
Discussion
Polymerase proofreading domain mutations can cause deficiency in exonuclease activity and thus result in increased mutation rate, which subsequently may confer the tumorigenesis (27). Besides, proofreading mutation in POLE are associated with better prognosis in endometrial cancer patients, who underwent adjuvant treatment of chemotherapy or radiotherapy following surgery (45,46).
By interrogating the genomic profiles of 21,074 Chinese cancer patients spanning 23 cancer types, we revealed a mutational prevalence of 3.2% for POLE and 1.4% for POLD1. Consistent with the results of the current study, Yao et al. demonstrated a mutation rate of 3.4% for POLE and 2.3% for POLD1 in 1,392 Chinese patients with lung, colorectum, liver, pancreatic, or stomach cancer (47). Another study that interrogated the mutational data of 47,721 patients from the cBioPortal database described mutation frequencies of 2.79% and 1.37% for POLE and POLD1, respectively, across multiple cancer types (22). We observed a comparable mutational frequency for POLE/POLD1 in the TCGA database (4.34%) versus BR VarDB (4.4%). Of note, TCGA tumors had a slightly higher frequency of co-occurring POLE and POLD1 mutations than our cohort (0.43% vs. 0.21%, P<0.001), which might be explained by differences in ethnicity or the differential distribution of histological subtypes.
Previous studies have reported controversial results regarding the association of POLE/POLD1 mutation with MSI status (13,47-49). In our study, POLE/POLD1-mutant tumors showed higher enrichment of genomic signatures correlated with POLE and MMR deficiencies compared with POLE/POLD1 WT tumors (Figure 3B). Genomic signatures for concurrent loss of polymerase proofreading and MMR function have been identified in POLE/POLD1-mutant endometrial cancer (50), which appeared in a non-additive manner and could represent the biological interaction of POLE/POLD1- and MMR-mediated DNA repair. It is possible that MSI could result in a mutation in POLE/POLD1 proofreading domain; or vice versa, the defective POLE/POLD1 proofreading function could cause a loss-of-function mutation in the MMR pathway, leading to MSI. If the former assumption is true, then we would expect POLE/POLD1 mutations to accumulate in MSI tumors as subclones; however, this conflicts with our observations in this study (Figure 4C). Moreover, we observed that while MSI frequently appeared in tumors harboring the dominant clone of the POLE/POLD1 mutation, it rarely occurred in POLE/POLD1 WT tumors or in tumors accumulating sub-cloned POLE/POLD1 mutations (Figure 4B). Collectively, our results indicate that POLE/POLD1 mutations are drivers of hypermutation, and MMR, leading to MSI, appears as the outcome in these tumors, which is in keeping with previous reports (30,50).
A recent study conducted in a pan-cancer cohort of 1,661 patients revealed POLE/POLD1 mutations to have a promising predictive value for positive outcomes after ICI treatment (22). However, Rousseau et al. questioned the results of this study due to the fact only a few hotspot mutations in or close to the proofreading domain of POLE/POLD1 had functional evidences (23). They also pointed out that the longer survival observed in POLE/POLD1-mutant patients was more than likely attributable to high TMB, given that the study mainly encompassed hypermutated tumors. Therefore, we carried out a multivariable Cox regression analysis on the same dataset with adjustment for factors including TMB (Figure 5B), and revealed that POLE/POLD1 mutation was not independently associated with OS. Interestingly, we found a significantly higher TMB in tumors harboring mutations in the proofreading domain versus those with mutations in other regions of POLE/POLD1 (Figure 4A). Together, our observations suggest that mutations in the proofreading domain of POLE/POLD1 are more likely to result in DNA repair defects and an extremely high TMB, which subsequently generate numerous neoantigens that contribute to sensitivity to ICI treatment (23,30). Advantage of POLE/POLD1 mutation as predictor.
POLE p.V411L and p. P286S/H/R are the most commonly found proofreading mutations associated with hypermutation in tumors (51). In our cohort, these mutations were identified respectively in two colorectal cancers and one ovarian cancer with extremely high TMB (155–450/Mb) (Table 2). We also uncovered two novel proofreading mutations, POLE p.Q292* and p.D406N, in a colorectal cancer and a gastric hypermutated cancer, respectively. Intriguingly, our study revealed a higher TMB in tumors with non-proofreading mutations than in POLD1/POLE WT tumors (Figure 4A), as well as a better ORR in POLE/POLD1-mutant patients, regardless of mutation location (Table 1). These observations are in agreement with the discovery of driver mutations in POLD1/POLE outside the exonuclease domain (30), suggesting that other domains may also function in proofreading. We also identified several mutations in the non-proofreading domain of POLE or POLD1 that potentially drive the hypermutation (Table 2). Among them, POLD1 p.E245K has been reported to be a driver mutation (30). Nonetheless, further functional experiments and clinical studies are required to verify the role of these newly identified mutations.
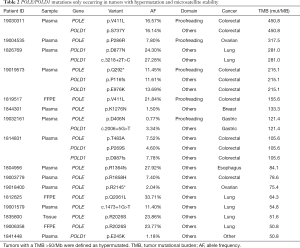
Full table
In this study, an elevated fraction of CD8+ TILs was observed in POLE/POLD1-mutant tumors, which is in accordance with what has been reported in POLE-mutant colorectal and endometrial cancers (17,52). van Gool et al. showed that POLE-mutant endometrial cancers harbored an increased number of CD8+ TILs and elevated expression of CD8A, accompanied by up-regulation of T cell exhaustion markers. Their findings indicated an enhanced cytotoxic T-cell response in POLE/POLD1-mutant cancers, which is probably attributable to a high neoantigen load (51,53).
In conclusion, our study investigated the prevalence and spectra of POLE and POLD1 mutations in 21,074 Chinese cancer patients and described differential mutational signatures in POLE/POLD1-mutated and WT cancers. Furthermore, POLE/POLD1 mutations were found to drive a higher TMB in tumors, with those occurring in the proofreading domain affecting the TMB more. Therefore, POLE/POLD1 mutations in the proofreading domain might have a prognostic value for cancer patients who receive ICI treatment. Our results also indicate that MSI caused by a loss-of-function mutation in the MMR pathway is the outcome of POLE/POLD1 proofreading deficiency in POLE/POLD1-mutant and MSI tumors.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/atm-20-7553
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-7553). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the institutional review board (IRB) of The First Affiliated Hospital of Zhengzhou University. Informed consent was not required due to the retrospective nature of the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Aarts BM, Klompenhouwer EG, Rice SL, et al. Cryoablation and immunotherapy: an overview of evidence on its synergy. Insights Imaging 2019;10:53. [Crossref] [PubMed]
- Ottaviano M, De Placido S, Ascierto PA. Recent success and limitations of immune checkpoint inhibitors for cancer: a lesson from melanoma. Virchows Arch 2019;474:421-32. [Crossref] [PubMed]
- Marin-Acevedo JA, Dholaria B, Soyano AE, et al. Next generation of immune checkpoint therapy in cancer: new developments and challenges. J Hematol Oncol 2018;11:39. [Crossref] [PubMed]
- Wang D, Lin J, Yang X, et al. Combination regimens with PD-1/PD-L1 immune checkpoint inhibitors for gastrointestinal malignancies. J Hematol Oncol 2019;12:42. [Crossref] [PubMed]
- Darvin P, Toor SM, Sasidharan Nair V, et al. Immune checkpoint inhibitors: recent progress and potential biomarkers. Exp Mol Med 2018;50:1-11. [Crossref] [PubMed]
- Yarchoan M, Hopkins A, Jaffee EM. Tumor Mutational Burden and Response Rate to PD-1 Inhibition. N Engl J Med 2017;377:2500-1. [Crossref] [PubMed]
- Tray N, Weber JS, Adams S. Predictive Biomarkers for Checkpoint Immunotherapy: Current Status and Challenges for Clinical Application. Cancer Immunol Res 2018;6:1122-8. [Crossref] [PubMed]
- Tremblay-LeMay R, Rastgoo N, Chang H. Modulating PD-L1 expression in multiple myeloma: an alternative strategy to target the PD-1/PD-L1 pathway. J Hematol Oncol 2018;11:46. [Crossref] [PubMed]
- Herbst RS, Soria JC, Kowanetz M, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014;515:563-7. [Crossref] [PubMed]
- Buchbinder EI, Desai A. CTLA-4 and PD-1 Pathways: Similarities, Differences, and Implications of Their Inhibition. Am J Clin Oncol 2016;39:98-106. [Crossref] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330-7. [Crossref]
- Cancer Genome Atlas Research Network; Kandoth C, Schultz N, et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013;497:67-73. [Crossref] [PubMed]
- Palles C, Cazier JB, Howarth KM, et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet 2013;45:136-44. [Crossref] [PubMed]
- Bellido F, Pineda M, Aiza G, et al. POLE and POLD1 mutations in 529 kindred with familial colorectal cancer and/or polyposis: review of reported cases and recommendations for genetic testing and surveillance. Genet Med 2016;18:325-32. [Crossref] [PubMed]
- Church DN, Briggs SE, Palles C, et al. DNA polymerase epsilon and delta exonuclease domain mutations in endometrial cancer. Hum Mol Genet 2013;22:2820-8. [Crossref] [PubMed]
- Shinbrot E, Henninger EE, Weinhold N, et al. Exonuclease mutations in DNA polymerase epsilon reveal replication strand specific mutation patterns and human origins of replication. Genome Res 2014;24:1740-50. [Crossref] [PubMed]
- Domingo E, Freeman-Mills L, Rayner E, et al. Somatic POLE proofreading domain mutation, immune response, and prognosis in colorectal cancer: a retrospective, pooled biomarker study. Lancet Gastroenterol Hepatol 2016;1:207-16. [Crossref] [PubMed]
- Mehnert JM, Panda A, Zhong H, et al. Immune activation and response to pembrolizumab in POLE-mutant endometrial cancer. J Clin Invest 2016;126:2334-40. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Gong J, Wang C, Lee PP, et al. Response to PD-1 Blockade in Microsatellite Stable Metastatic Colorectal Cancer Harboring a POLE Mutation. J Natl Compr Canc Netw 2017;15:142-7. [Crossref] [PubMed]
- Zhu B, Liu Y, Li J, et al. Exceptional Response of Cryoablation Followed by Pembrolizumab in a Patient with Metastatic Cervical Carcinosarcoma with High Tumor Mutational Burden: A Case Report. Oncologist 2020;25:15-8. [Crossref] [PubMed]
- Wang F, Zhao Q, Wang YN, et al. Evaluation of POLE and POLD1 Mutations as Biomarkers for Immunotherapy Outcomes Across Multiple Cancer Types. JAMA Oncol 2019;5:1504-6. [Crossref] [PubMed]
- Rousseau B, Vidal J, Diaz LA Jr. Evaluation of POLE/POLD1 Variants as Potential Biomarkers for Immune Checkpoint Inhibitor Treatment Outcomes. JAMA Oncol 2020;6:589-90. [Crossref] [PubMed]
- Vilar E, Gruber SB. Microsatellite instability in colorectal cancer-the stable evidence. Nat Rev Clin Oncol 2010;7:153-62. [Crossref] [PubMed]
- Dudley JC, Lin MT, Le DT, et al. Microsatellite Instability as a Biomarker for PD-1 Blockade. Clin Cancer Res 2016;22:813-20. [Crossref] [PubMed]
- Henninger EE, Pursell ZF. DNA polymerase epsilon and its roles in genome stability. IUBMB Life 2014;66:339-51. [Crossref] [PubMed]
- Rayner E, van Gool IC, Palles C, et al. A panoply of errors: polymerase proofreading domain mutations in cancer. Nat Rev Cancer 2016;16:71-81. [Crossref] [PubMed]
- Albertson TM, Ogawa M, Bugni JM, et al. DNA polymerase epsilon and delta proofreading suppress discrete mutator and cancer phenotypes in mice. Proc Natl Acad Sci U S A 2009;106:17101-4. [Crossref] [PubMed]
- Herr AJ, Kennedy SR, Knowels GM, et al. DNA replication error-induced extinction of diploid yeast. Genetics 2014;196:677-91. [Crossref] [PubMed]
- Campbell BB, Light N, Fabrizio D, et al. Comprehensive Analysis of Hypermutation in Human Cancer. Cell 2017;171:1042-56 e10.
- Billingsley CC, Cohn DE, Mutch DG, et al. Polymerase varepsilon (POLE) mutations in endometrial cancer: clinical outcomes and implications for Lynch syndrome testing. Cancer 2015;121:386-94. [Crossref] [PubMed]
- Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019;51:202-6. [Crossref] [PubMed]
- Miao D, Margolis CA, Vokes NI, et al. Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat Genet 2018;50:1271-81. [Crossref] [PubMed]
- Gandara DR, Paul SM, Kowanetz M, et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med 2018;24:1441-8. [Crossref] [PubMed]
- Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015;350:207-11. [Crossref] [PubMed]
- Hugo W, Zaretsky JM, Sun L, et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell 2016;165:35-44. [Crossref] [PubMed]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754-60. [Crossref] [PubMed]
- McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297-303. [Crossref] [PubMed]
- Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012;22:568-76. [Crossref] [PubMed]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [Crossref] [PubMed]
- Cingolani P, Platts A. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80-92. [Crossref] [PubMed]
- Newman AM, Bratman SV, Stehr H, et al. FACTERA: a practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics 2014;30:3390-3. [Crossref] [PubMed]
- Diaz-Gay M, Vila-Casadesus M, Franch-Exposito S, et al. Mutational Signatures in Cancer (MuSiCa): a web application to implement mutational signatures analysis in cancer samples. BMC Bioinformatics 2018;19:224. [Crossref] [PubMed]
- Zhu L, Huang Y, Fang X, et al. A Novel and Reliable Method to Detect Microsatellite Instability in Colorectal Cancer by Next-Generation Sequencing. J Mol Diagn 2018;20:225-31. [Crossref] [PubMed]
- Stelloo E, Bosse T, Nout RA, et al. Refining prognosis and identifying targetable pathways for high-risk endometrial cancer; a TransPORTEC initiative. Mod Pathol 2015;28:836-44. [Crossref] [PubMed]
- Church DN, Stelloo E, Nout RA, et al. Prognostic significance of POLE proofreading mutations in endometrial cancer. J Natl Cancer Inst 2015;107:402. [Crossref] [PubMed]
- Yao J, Gong Y, Zhao W, et al. Comprehensive analysis of POLE and POLD1 Gene Variations identifies cancer patients potentially benefit from immunotherapy in Chinese population. Sci Rep 2019;9:15767. [Crossref] [PubMed]
- Wong A, Kuick CH, Wong WL, et al. Mutation spectrum of POLE and POLD1 mutations in South East Asian women presenting with grade 3 endometrioid endometrial carcinomas. Gynecol Oncol 2016;141:113-20. [Crossref] [PubMed]
- Jansen AM, van Wezel T, van den Akker BE, et al. Combined mismatch repair and POLE/POLD1 defects explain unresolved suspected Lynch syndrome cancers. Eur J Hum Genet 2016;24:1089-92. [Crossref] [PubMed]
- Haradhvala NJ, Kim J, Maruvka YE, et al. Distinct mutational signatures characterize concurrent loss of polymerase proofreading and mismatch repair. Nat Commun 2018;9:1746. [Crossref] [PubMed]
- Nebot-Bral L, Brandao D, Verlingue L, et al. Hypermutated tumours in the era of immunotherapy: The paradigm of personalised medicine. Eur J Cancer 2017;84:290-303. [Crossref] [PubMed]
- Howitt BE, Shukla SA, Sholl LM, et al. Association of Polymerase e-Mutated and Microsatellite-Instable Endometrial Cancers With Neoantigen Load, Number of Tumor-Infiltrating Lymphocytes, and Expression of PD-1 and PD-L1. JAMA Oncol 2015;1:1319-23. [Crossref] [PubMed]
- van Gool IC, Eggink FA, Freeman-Mills L, et al. POLE Proofreading Mutations Elicit an Antitumor Immune Response in Endometrial Cancer. Clin Cancer Res 2015;21:3347-55. [Crossref] [PubMed]
(English Language Editor: J. Reynolds)


