A narrative review of the roles of the miR-15/107 family in heart disease: lessons and prospects for heart disease
Introduction
MicroRNAs (miRNAs) are a class of small non-coding RNAs, generally 20–22 nucleotides(nt) in length. Most miRNAs post-transcriptionally inhibit target genes by binding to the 3’ untranslated region (3’UTR) of target genes, leading to the messenger RNA (mRNA) degradation or protein translation inhibition. Thus, miRNAs play a significant role in various biological processes, such as apoptosis, autophagy, and angiogenesis (1-3).
A miRNA family is defined as a group of miRNAs that possess seed sequence. The seed region usually refers to the 2–7 nt of the 5’ portion of miRNAs, which is critical for target recognition, and determines the overlapping target genes (4-6).
Heart diseases are multifactorial disorders, comprising myocardial infarction (MI), cardiac hypertrophy (CH), fibrosis, heart failure (HF), and other conditions. The morbidity and mortality of heart disease are increasing globally and greatly burden the individual and their family. Although a number of regimens are used to treat heart failure, its morbidity and mortality continue to rise. Therefore, efforts to develop new therapeutic strategies are required (7,8).
Numerous studies have demonstrated the regulatory role of miRNAs, such as miR-1, miR-22, and miR-133, in the development and progression of heart disease, and have provided us with a comprehensive overview of the role of miRNAs in cardiac development (9-11). Given the miR-15/107 family members harbor identical seed regions—which means there is a high degree of overlap in target genes—this review focuses on the miR-15/107 family and summarizes their physiological and pathological effects on heart disease, their circulating signature in patients with heart disease, and the current applications and challenges of the miR-15/107 family in clinical trials. By conducting this review, we hope to provide guidance for the diagnosis, prognosis, and treatment of heart disease.
We present the following article in accordance with the Narrative Review reporting checklist (available at http://dx.doi.org/10.21037/atm- 20-6073).
Methods
We conducted a literature search of the PubMed database using the following keywords: “miR-15/107 family”, specific individual miRNAs of the miR-15/107 family and heart disease, “circulating miR-15/107 family and heart disease”. We also reviewed articles on the concepts of “miRNA”, “miRNA family”, and “miRNA cluster”, as well as the definition of different heart diseases, including “myocardial hypertrophy”, “myocardial infarction”, “diabetic cardiomyopathy”, “hypertensive heart disease”, and “heart failure”. Original articles and review articles published in the English language between April 2002 to May 2020 were included.
Genomics location, conservation, and expression pattern of miR-15/107 family
The miR-15 family consists of 6 paralogous members: miR-15a, miR-15b, miR-16, miR-195, miR-424, and miR-497. Based on the conserved “AGCAGC” sequence, the miR-15 family may be extended to include the miR-15/107 family, which comprises miR-103, miR-107, miR-503, and miR-646 (12).
The miR-15/107 family is ubiquitously expressed in different tissues and evolutionarily conserved; however, miR-497, miR-195, miR-424, and miR-503 are only expressed in mammals, while miR-646 is human- and chimpanzee-specific (12). The miR-15/107 family is expressed at medium-to-high levels across various tissues. While the family members are ubiquitous and not tissue specific, they show tissue preference. For instance, miR-103 and miR-107 are expressed at higher levels in the brain, while other family members are expressed at higher levels in non-brain tissue. Furthermore, miR-15a, miR-15b, and miR-16 show higher expression in the spleen, while miR-424 is enriched in the kidney, liver, and skeletal muscle. Kidney tissue also has a relatively high expression of miR-646 (13). Only miR-103 and miR-107 are included on the list of the 18 miRNAs constitutively expressed in all mammalian tissues during different stages of development (14). miR-15b and miR-16 are upregulated in the late stages of erythropoiesis (15,16), while the expression levels of miR-15 and miR-16 are relatively higher in the fetal brain (17).
The miR-15/107 family members can be classified into different clusters according to their chromosomal locations. miRNAs in the same clusters tend to function similarly with similar transcriptional regulations (18). The miR-15a and miR-16-1 cluster is located at the deleted in lymphocytic leukemia 2 (DLEU2) gene on chromosome 13. The miR-15b and miR-16-2 cluster is situated in the structural maintenance of chromosomes 4 (SMC4) gene. SMC4 is a critical, highly conserved ATPase that involved in DNA/chromatin dynamics. miR-16-1 and miR-16-2 are transcribed from DLEU2 and SMC4, respectively, and progress to the same mature miR-16 sequence. Specifically, miR-15a and miR-16 are located within the fragile region of chromosome 13q, which is deleted in chronic lymphocytic leukemia (CLL) (19). The miR-497 and miR-195 cluster is derived from a long non-coding RNA, MIR497HG, which resides in chromosome 17p13.1. The miR-424 and miR-503 cluster is located at the same locus of chromosome X. Notably, miR-322 is the homolog of miR-424 in mouse (12,20). Despite miR-103 and miR-107 being located on different chromosomes, their host genes belong to the pantothenate kinase (PANK) family (21). miR-103 and miR-107 are almost identical but differ by one nucleotide at position 21 (22), and recent studies have revealed their cooperative role in various diseases. Unlike the miR-15 family, the seed sequence of miR-103/107 starts from the first nucleotide of the 5’ end (22). Consequently, in the prediction of target genes using TargetScan (http://www.targetscan.org/vert_71/), the predicted target genes of the miR-15 family differ considerably from those of miR-103/107.
Since the miR-15/107 family harbors the same seed sequence, the target genes of these miRNAs overlap (23) and thus regulate a series of biological processes including proliferation, cell division, metabolism, stress response, and angiogenesis (12). Additionally, miR-15/107 can regulate target genes by binding preferentially to their coding sequence (CDS) region (24). Dysregulation of miR-15/107 family members has been reported in various disease, including cardiovascular diseases, neurodegenerative disease, and cancer (12).
The evolution of the miR-15/107 family has been investigated; however, whether members of miR-15/107 family descend from the same or different ancestors has not been studies and requires further investigation.
Function of the miR-15/107 family in heart development and cardiac disease
Cheng et al. performed mouse miRNA microarray analysis and revealed a high abundance of miR-15a, miR-16, miR-424, miR-195, and miR-103 in normal mouse hearts tissues, suggesting that the potential importance of this miRNA family (25). This research has prompted further exploration of the miR-15/107 family in cardiac function.
Studies have demonstrated that the miR-15/107 family members not only participate in the development of the heart but are dysregulated in heart diseases, including in the condition of myocardial hypertrophy, myocardial infarction, diabetic cardiomyopathy, hypertensive heart disease, and heart failure.
miR-195 in different types of heart disease
Currently, miR-195 is the most extensively studied miRNAs in relation to cardiac function. Here, we discuss the involvement of miR-195 in different heart diseases, including myocardial hypertrophy, myocardial infarction, diabetic cardiomyopathy, and heart failure.
miR-195 in postnatal cardiomyocyte mitotic arrest
Post-natal cardiomyocyte mitotic arrest significantly limits the regenerative ability of the adult mammalian heart after injury (26). In the first study of illuminating the role of the miR-15 family in heart development, Porrello et al. defined the miR-15 family members as significant regulators of post-natal cardiomyocyte mitotic arrest (27). During the neonatal period, members of the miR-15 family, especially miR-195, are upregulated, accounting for cardiomyocyte mitotic arrest. Further analysis demonstrated that miR-195 downregulated cell cycle genes, including Cdc2a, Chek1, Birc5, Nusap1, and Spag5. In contrast, silencing of the miR-15 family upregulates cell cycle genes, inducing cardiomyocyte mitotic entry and progression during the post-natal period (27).
miR-195 in myocardial hypertrophy
Myocardial hypertrophy is a compensatory response of the heart to increase pressure overload. Without effective intervention, persistent myocardial hypertrophy leads to decompensation of cardiac function before eventual progression to heart failure. Therefore, elucidating the underlying mechanism of the development and progression of myocardial hypertrophy may guide new treatments for heart failure management (28).
Upregulation of miR-195 has consistently been reported in different models of myocardial hypertrophy, including thoracic aortic banding (TAB)-induced (29), calcineurin A (CnA)-induced (29), isoprenaline (ISO)-induced (30), and angiotensin 2 (Ang2)-induced hypertrophic models (31,32). Expression of miR-195 is also induced in human failing myocardium (33); however, the mechanism by which miR-195 regulates myocardial hypertrophy vaires. miR-195 inhibits myocardial hypertrophy mainly by regulating cell cycle-related genes (34), mitochondrial integrity and function (32), energy metabolism (33), and the TGFβ signaling pathway (31). Cell-cycle regulator cyclin D2 (34), HMGA1 (30), mitofusin 2 (MFN2) (32), FBXW7 (32), and SIRT3 (33) have been confirmed as be target genes of miR-195 involved in these biological events. Of note, MFN2 is a mitochondrial outer membrane GTPase that mediates mitochondrial clustering and fusion (35), and modulates the generation of reactive oxygen species (ROS) (36,37). Deceased SIRT3 expression can also be found in failing human myocardium. As a mitochondrial deacetylase, SIRT3 regulates mitochondrial acetylome and maintains mitochondrial function (33). Mitochondrial disruption has a crucial role in the pathophysiological process of heart disease (8,38); hence, it is interesting to further explore the regulation of mitochondrial integrity and function by miR-195.
Finally, in mouse models of hypertrophic cardiomyopathy, miR-195 has been shown to be enriched, resulting in the inhibition of Cab39 and repression of the Lkb1/Strad/Cab39-dependent signaling pathway (39).
miR-195 in myocardial infarction
In myocardial infarction (MI), miR-195 is also found to be elevated. MI refers to myocardial cell death due to prolonged ischemia (40). Apoptosis, also known as programmed cell death, is major forms of cardiomyocyte death during MI (41). Oxygen deprivation or an increase oxidative stress can trigger cell apoptosis and evoke MI (38).
miR-195 has been found to be consistently upregulated in various induced models of MI, such as ischemia reperfusion(I/R) injury in mice (42), MI in rats (43), hypoxia/reoxygenation (H/R) treatment of H9C2 cells (44), hypoxic (43) and H2O2 treatment (43,45) of cardiomyocytes, as well as palmitate-induced myocardial apoptosis (46). In these different injuries, miR-195 predominantly functions mainly by targeting a series of anti-apoptotic target genes, such as BCL2 (42,43), c-myb (44), BCL2L2 (45), and SIRT1 (46), to induce ROS generation and promote apoptosis. In particular, c-myb suppresses mitochondrial superoxide generation (44), which supports the role of miR-195 in mitochondrial homeostasis. The findings above highlight the pro-apoptotic role of miR-195 and also encourage further investigation into interactions in the miR-195/target genes/apoptosis axis.
Porrello et al. uncovered the potential role of the miR-15 family in heart regeneration (27). Follow-up studies by this group established ischemic MI models to further illuminate the cardiac regenerative capacity of miR-15 family members, and confirmed that the family mediates neonatal heart regeneration via suppression of postnatal cardiomyocyte proliferation. Moreover, hearts with transgenic miR-195 were found to have lower regenerative capacity after MI, while suppression of miR-15 family members led to adult myocyte proliferation and improved cardiac function after MI. Porrello et al. also demonstrated that the miR-15 family suppresses cardiomyocyte proliferation and a number of cell cycle-related genes (47).
miR-195 in diabetic cardiomyopathy
Diabetic cardiomyopathy is a cardiac disease associated with diabetes (48). In a mouse model of streptozotocin (STZ)-induced Type 1 diabetes, miR-195 was observed to be upregulated (49); however, in a rat model of high-fat high-fructose diet-induced Type 2 diabetes, miR-195 was downregulated (50). BCL2 and SIRT1 were characterized as target genes of miR-195 in both these models (49,50). Zheng et al. further reported that overexpression of miR-195 induced apoptosis and inhibited angiogenesis, while suppression of miR-195 reduced oxidative damage and promoted angiogenesis (49). Moreover, Khakdan et al. demonstrated that high-intensity interval training (HIIT) can effectively improve cardiac function via downregulation miR-195 (50).
miR-195 in heart failure
Heart failure is the end-stage of most heart disease, including myocardial hypertrophy, hypertension heart disease, and MI (51,52). In a mouse model of transverse aortic constriction (TAC)-induced heart failure, inhibition of miR-195 was found to mitigate heart function injury in heart failure by targeting CXCR4 and inactivating the JAK/STAT signaling pathway (53). Xie et al. reported that miR-195 was downregulated in cardiac disease. In rats with heart failure, the myocardium showed a lower expression of miR-195 and a higher protein expression of TGF-β1 and SMAD3. Enhanced expression of miR-195 can repress H/R-induced cardiomyocyte apoptosis and thus preserve cardiac function (54); however, the mechanism underlying this inconsistency remains elusive. Furthermore, miR-195 is upregulated in cardiac dysfunction induced by selenium deficiency (55).
These findings show that miR-195 is mostly upregulated in different settings of heart disease and behaves as a detrimental cardiac miRNA under different injuries.
In addition to miR-195, both in vivo and in vitro models were established by multiple independent research groups to extend the understanding of the role of other miR-15/107 family members in the cardiovascular system.
miR-497 in different types of heart disease
Despite miR-497 and miR-195 belonging to the same cluster, their functions in cardiac development and disease are quite different.
miR-497 is decreased during cardiac differentiation (56), as well as in models of Ang2- and TAC-induced hypertrophy (57). During the induction of cardiac differentiation, miR-497 targets TGFβ1, TGFβR1, TGFβR2, and SMAD3, and regulates the TGFβ signaling pathway (56). SIRT4 has been identified as a target of miR-497 in hypertrophic inhibition (57). Specifically, SIRT4 is a mitochondrial NAD+-dependent ADP-ribosyltransferase, that participates in metabolism (58,59). This supports that the miR-15/107 family has critical involvement in mitochondrial regulation and energy metabolism.
The expression patterns of miR-497 in response to injury are contradictory. Li et al. defined miR-497 as a harmful modulator in neonatal rat cardiomyocytes (NRCs) with anoxia/reoxygenation injury, as well as in murine hearts subjected to MI injury. During injury, miR-497 is decreased. Overexpression of miR-497 induces apoptosis and suppresses autophagy by inhibiting BCL2 and autophagy related gene light chain 3 B (LC3B) (60); however, in mouse models of myocardial I/R injury, overexpression of miR-497 has been reported to downregulate MFN2, thus inhibiting cardiomyocytes apoptosis (61).
miR-424 in different types of heart disease
In a study to identify miRNAs expressed in early cardiac progenitors cells, the miR-322 (ortholog of human miR-424) and miR-503 cluster was found to be expressed in the early stage of development of cardiac progenitor cells and to regulate cardiac muscle differentiation during embryogenesis, which further evidenced to the role of the miR-15/107 family in cardiac development (20).
Studies from different groups have demonstrated that the level of miR-424 increases with hypoxia (62,63). The upregulation of miR-424 results in the accumulation of HIF1α in the nucleus, which enhances angiogenesis (62). Overexpression of miR-424 is HIF1α-dependent, and leads to VEGF reduction and the enhancement of vascular endothelial cadherin phosphorylation (63).
miR-424 is also increased in cardiac I/R damage. In a mouse model of I/R injury and cell models subjected to H/R injury, miR-424 was shown to be upregulated, and triggered the overexpression of several pyroptosis-related proteins, such as caspase-1, caspase 11, IL-1β, and IL8 by targeting CRISPLD2 (64).
miR-16 in different types of heart disease
miR-16 behaves as a protective factor in cardiac hypertrophy. Huang et al. established different animal and cell models to illustrate the contribution of miR-16 in cardiac hypertrophy. miR-16 has consistently been found downregulated in rat models with abdominal aortic constriction (AAC), mouse models subjected to TAC, mouse models induced by phenylephrine (PE), and cells treated with PE or Ang2. Mechanistically, overexpression of miR-16 inhibits cardiac hypertrophy by targeting cyclins D1, D2, and E1. Conversely, silencing of miR-16 results in activation of the cyclin/Rb pathway and promotes progression to cardiac hypertrophy (65).
In contrast to its cardioprotective role in cardiac hypertrophy, miR-16 has been identified as having an adverse effect on the pathogenesis of ischemic heart disease. Expression of miR-16 is induced in H/R-treated AC16 cells (66), H2O2-treated neonatal rat ventricular cells (NRVCs), and rats with acute myocardial infarction (AMI) (67). Moreover, miR-16 aggravates cardiac injury by decreasing cell viability and increasing apoptosis (66,67). IRS1 (66) and beta2-adrenoreceptor (β2-AR) (67) are experimentally established targets of miR-16. These studies demonstrates that miR-16 is involved in MI.
In hypertensive heart disease (HHD), another cause of heart failure (68), miR-16 expression is also increased (69). He et al. aimed to explore the mechanism underlying this, and demonstrated that miR-16 negatively regulates the adrenoreceptor alpha 1a gene (ADRA1A) in deoxycorticosterone acetate (DOCA)-induced cell and rat models of HHD (70).
miR-15b in different types of heart disease
Several studies have determined that miR-15b is upregulated in MI (71-74). BCL2, ARL2, PDK4, and SGK1 have been identified as target genes of miR-15b during injury (74-76). Notably, PDK4 is involved in mitochondrial function (77), SGK1 is an anti-apoptosis protein (78), and ARL2 is a regulator of mitochondrial integrity and cardiac function (76). Given the role of mitochondrial dysfunction in heart disease, it is likely that disruption of miR-15b degenerates the mitochondrial by controlling ATP production as well as ROS formation, which ultimately culminates in heart disease.
Furthermore, miR-15b is elevated in hypertrophic cardiomyopathy, by targeting p38, TGFBR1, TGFBR2, TGFBR3, SMAD3, SMAD7, and regulating the TGFβ signaling pathway (79), which is similar to the regulation of cardiac differentiation by miR-497 (56).
Conversely, Liu et al. reported reduced expression of miR-15b in human umbilical vein endothelial cells (HUVECs) during hypoxia. Hypoxia-induced tube formation and cell migration were attenuated after transfection with miR-15b, and were accompanied by the downregulation of VEGF and Ang2, suggesting the possible involvement of miR-15b in anti-angiogenic activity (80).
miR-103/107 in different types of heart disease
miR-103 and miR-107 are defined as cardioprotective factors in cardiac hypertrophy. Rat models with TAC and Ang2-treated cells showed a reduction of miR-103 expression, along with a concurrent upregulation of TRPV3 and increased autophagy. Furthermore, overexpression of miR-103 reduced BNP and β-MHC expression, and reduced fluorescence intensity of Ca2+ signal, indicating the inhibitory effect of miR-103 on cardiac hypertrophy (81). Rech et al. further demonstrated that inhibition of miR-103/107 attenuated cardiac function, leading to decrease in cardiomyocyte size and mitochondrial oxidative capacity (82). Since miR-103 and miR-107 have reached the clinical stage in the treatment of patients with Type 2 diabetes and liver disease on account of their upregulation in obese mice and insulin sensitivity (83), their protective role in cardiac function emphasizes that accurate therapeutic delivery of drugs to specific organs is imperative to avoiding side effects (82).
Both in vivo and in vitro studies have demonstrated the positive involvement of miR-103/107 in necrosis. Exposure to high doses of H2O2 leads to a reduction of H19, which releases miR-103/107, thereby reducing FADD, which negatively affects cell necrosis by inhibiting the formation of the PIPK1/PIPK3 complex. Accordingly, suppression of miR-103/107 can attenuate necrosis (84). Wang et al. also revealed that miR-103/107 show no effect on TNF-α-induced necrosis and low concentration of H2O2-initiated apoptosis, indicating that the phenotypic effects of the same molecule may differ depending on the type and dose of stress (84).
The entire miR-15/107 family in different types of heart disease
Given that the miR-15/107 family shares an identical seed sequence, some researchers have also studied the family as a whole. Data from Sayed et al. reveals that miR-15b, miR-195, miR-103, and miR-107 were upregulated 14 days after TAC (85). Tijsen et al. showed that miR-15b, miR-16, miR-497, miR-195, and miR-424 were consistently increased in hypertrophic cardiomyopathy (79), thus confirming the involvement of the miR-15/107 family in cardiac hypertrophy.
Consistently elevated levels of miR-15a, miR-15b, miR-16, miR-497, and miR-195 were detected in the infarcted zone 24 hours after ischemic injury in a porcine MI models study (74), which evidenced the role of the miR-15/107 family in ischemic heart disease.
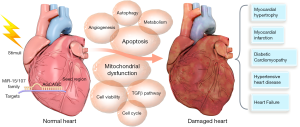
Here, we have summarized the expression patterns of the miR-15/107 family members in different models of heart damage (Table 1) and the possible mechanisms leading to heart damage (Figure 1).

Full table
Circulating miR-15/107 family members as biomarkers for patients with heart disease
miRNAs are initially discovered circulating in the blood. Circulating miRNAs possess great potential as biomarkers for cancer and heart disease due to their abundance, high stability in the blood, high specificity, and strong diagnostic sensitivity (86-88).
Although troponin and creatine kinase—the current gold standard for diagnosis for MI and heart failure—show strong sensitivity, their specificity is somehow insufficient, as troponin and creatine kinase are also elevated in patients with chronic renal disease (89). Searches for alternative biomarkers with higher specificity and sensitivity for heart disease are urgently needed.
miR-195 was found to be increased in plasma samples from patients with AMI compared to healthy individuals, suggesting that it could be a potential biomarker for AMI (90). Other members of the miR-15/107 family also circulate miRNAs in the blood of patients with heart disease.
For instance, miR-16 is significantly increased in the plasma of patients with takotsubo cardiomyopathy (TTC) compared to healthy individuals. This unique signature of elevated circulating miR-16-5p also distinguishes TTC from MI patients (91).
Furthermore, miR-16 is also upregulated in the urine and serum of cardiac surgery patients, as well as being higher in the urine of on-pump patients than off-pump patients. This suggests that the release of specific miRNAs is likely to be responsible for ischemic and hypoxic damage during surgery (92).
Interestingly, the signature of circulating miRNAs signature differs in patients with heart failure depending on the type and stage of disease. Plasma samples from patients with acute heart failure display a reduction in the level of miR-103 (93). Another study showed that miR-16 is negatively correlated with C-reactive protein, which indicates a worse clinical outcome for patients with acute heart failure (94). The peripheral blood mononuclear cells (PBMC) of patients with chronic heart failure show a downregulation of miR-497 and miR-107 (95), while miR-16 is decreased in the peripheral blood of patients with end-stage heart failure. And vacuolar protein sorting 4a (VPS4a), an ATPase that significantly decreases total cell numbers, has been identified as a target of miR-16, and it is increased during end stage of heart failure patients. Moreover, after left ventricular assist device (LVAD) therapy, miR-16 is increased, and VPS4a is decreased (96). Consistently, in silico analysis of the peripheral blood samples of patients with heart failure revealed that miR-16, miR-424, and miR-195 were decreased in coronary sinus blood samples (97); however, data from Zhang et al. confirmed that miR-195 levels in the heart and plasma of patients with heart failure are increased (33). As previously mentioned, miR-16 is increased in HHD (70), it has also been found to be increased in the plasma of patients with hypertension-induced heart failure (69). Fourteen miRNAs including miR-15a are reported to be upregulated in patients with hypertrophic cardiomyopathy (98). Overall, the miRNA expression profiles in blood samples from patients with different types and stages of heart failure show diverse expression patterns of miR-15/107 family members. Whether this signature can be used to stratify heart disease also deserves deeper exploration.
Nevertheless, miRNAs levels are influenced by the selection of reference genes, the method of RNA extraction and analysis, the samples types (serum, plasma, or whole blood), and whether patients are hospitalized or undergoing drug treatment. Therefore, an industry-accepted standard is significant to accelerate the application of circulating miRNA in the clinical settings.
Given that the ‘area under the ROC curve’ (AUC) of a combination of several genes is higher than that of an individual gene (99), the signature of circulating miRNAs in patients with heart diseases may open new avenues for diagnosis through combining both protein and RNA biomarkers.
Collectively, the studies discussed above characterize the miR-15/107 family members as promising candidates for diagnostic, prognostic, and therapeutic application in heart disease.
We have now summarized the expression patterns of circulating miR-15/107 family in patients with different types of heart disease (Table 2).

Full table
Clinical applications and challenges of the miR-15/107 family in disease
Locked nucleic acid (LNA)-modified anti-miRs have been demonstrated to silence miRNAs efficaciously in vivo and show excellent potential for the treatment of diseases. For example, miR-122 inhibitor has been used to cure HCV infection (100,101).
It is reported that miRNAs have promising applicability in the treatment of heart disease. miR-208a, for instance, has been shown to play a crucial role in the development of cardiac hypertrophy and myocardial fibrosis (102), and the antagomiR of miR-208a: MGN-9103 has reached preclinical testing.
Inhibition of members of the miR-15/107 family has reached clinical trials for noncardiac disease. For example, administration of miR-16 is under investigation in patients with malignant pleural mesothelioma and non-small cell lung cancer (ClinicalTrials.gov Identifier: NCT02369198). Additionally, antagomiR-103/107 (known as RG-125/AZD4076) has gained a clinical phase I and IIA trial as a potential treatment for patients with Type 2 diabetes with non-alcoholic fatty liver disease (NAFLD) and non-alcoholic steatohepatitis (NASH) (ClinicalTrials.gov Identifiers: NCT02826525 and NCT02612662).
Given that members of the miR-15/107 family are dysregulated in various heart diseases, manipulation of members of the miR-15/107 family can attenuate the effects caused by various damage. Therefore, anti-miRs of the miR-15/107 family show excellent diagnostic, prognostic, and therapeutic potential for patients with heart disease, and deserves deeper investigation.
Discussion
This review of the collective work of independent research groups using both animal and cell models have illuminated the role of the miR-15/107 family in cardiac function. It is intriguing that the expression patterns of members of the same miRNA family are highly diverse. miRNAs in the same family may display opposite effects, and even the phenotypic effects of a single miRNA can differ under different conditions. We discovered that the contradictory phenotypic effects of miRNAs in the same family depend on the type and degree of stress, the timing of injury, and cell types and species. For example, miR-16 has a protective effect in cardiac hypertrophy, but has adverse effects in ischemic heart disease. miR-195 behaves consistently as a deleterious cardio-miRNA, while inhibition of miR-103/107 deteriorates the cardiac phenotype. The reason behind these differences remains unclear. We speculate that the following may be some potential reasons to explain these conflicting results: (I) different cell populations, different gene expression profiles; (II) the affinity between miRNAs and target genes; (III) the polymorphism in miRNA and miRNA target sites; (IV) the genomic location, which means that different miRNAs possess different neighboring genes; and (V) the upstream transcriptional factor of miRNAs. There is a lot of work left to carry out in order to address these perplexing phenomena.
Most studies to date have focused on individual miRNAs in the miR-15/107 family instead of the whole family. The effect of a single miRNA is often mild, and it has been widely recognized that a single miRNA can govern numerous target genes, conversely, a single target gene can be manipulated by multiple miRNAs (103,104). Thus, future studies are required to decipher the cooperation of the entire group. Co-regulation networks between miRNAs-targets are also needed to tackle the complexity of regulation. The ability of the entire family to outperform an individual miRNAs requires further investigation.
The results described above also demonstrate that targets of the miR-15/107 family involved in heart disease are most likely to be correlated with mitochondrial dysfunction and apoptosis. For example, SITR1, SIRT3, SIRT4, MFN2, ARL2, PDK4, and c-Myb are involved in mitochondrial regulation, and BCL2 and SGK1 are crucial in anti-apoptotic events. Based on the computational prediction using starbase.com (http://starbase.sysu.edu.cn/index.php) and TargetScan.com (http://www.targetscan.org/vert_71/), there are still a variety of predicted targets associated with mitochondria which remain to be experimentally validated. Given that mitochondrial dysfunction is critical for apoptosis and heart disorder (8,38), it is reasonable to speculate that the miR-15/107 family predominantly functions in heart disease mainly by mediating mitochondrial function and apoptosis. The involvement of other targets in the regulatory network involving the miR-15/107 family, mitochondrial, apoptosis, and heart disease, also requires further investigation. Since mitochondria-targeted antioxidant therapies have gained increasing attention, interventions to reserve mitochondrial dysfunction may offer a practical strategy to ameliorate heart disease. Therefore, deeper exploration of the interaction between miR-15/107 family members and mitochondrial may provide insights into the progression of heart disease. Finally, given the contradictory role of the miR-15/107 family in different diseases, particular attention should be paid to the hurdle posed by off-target effects in which an individual miRNA functions divergently by binding to distinct target genes. Consequently, it may be favorable to target cell-specific or tissue-specific downstream effectors, especially the mitochondrial-related targets instead of miRNAs themselves, in order to avoid side effects.
Conclusions
Numerous therapeutic approaches have been used in recent decades to repair heart damage and preserve cardiac function, however, there are still many knowledge gaps regarding the cellular and molecular mechanisms that mediate cardiac function, and these require further investigation.
In this review, both animal and cell models of heart disease have demonstrated disruption of the miR-15/107 family in response to diverse types of stress. Disruption of the miR-15/107 family promotes cardiac phenotype deterioration and plays an under-appreciated role in the development and progression of heart disorders. It has been shown that a collection of genes involved in proliferation, apoptosis, autophagy, energy metabolism, angiogenesis, and cell cycle are controlled by the miR-15/107 family. The current clinical application of the miR-15/107 family in noncardiac disease, as well as the circulating miR-15/107 family signature in patients with different heart diseases, indicates their compelling potential for clinical application in a cardiac population. Elucidating the different expression patterns and underlying mechanism of the miR-15/107 family will pave the way for attractive therapeutic applications.
Acknowledgments
Funding: This work was supported by Science and Technology Planning Project of Guangdong Province, China (2019B030316024), National Key Research and Development Project (2016YFC1300600), National Natural Science Foundation of China (U1601219, 81970420, 91939109).
Footnote
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-6073
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-6073). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [Crossref] [PubMed]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009;136:642-55. [Crossref] [PubMed]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 2009;10:126-39. [Crossref] [PubMed]
- Mourelatos Z. Small RNAs: The seeds of silence. Nature 2008;455:44-5. [Crossref] [PubMed]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005;120:15-20. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215-33. [Crossref] [PubMed]
- Ordovas JM, Smith CE. Epigenetics and cardiovascular disease. Nat Rev Cardiol 2010;7:510-9. [Crossref] [PubMed]
- Bonora M, Wieckowski MR, Sinclair DA, et al. Targeting mitochondria for cardiovascular disorders: therapeutic potential and obstacles. Nat Rev Cardiol 2019;16:33-55. [Crossref] [PubMed]
- Espinoza-Lewis RA, Wang DZ. MicroRNAs in heart development. Curr Top Dev Biol 2012;100:279-317. [Crossref] [PubMed]
- Zhu H, Fan GC. Role of microRNAs in the reperfused myocardium towards post-infarct remodelling. Cardiovasc Res 2012;94:284-92. [Crossref] [PubMed]
- Topkara VK, Mann DL. Role of microRNAs in cardiac remodeling and heart failure. Cardiovasc Drugs Ther 2011;25:171-82. [Crossref] [PubMed]
- Finnerty JR, Wang WX, Hébert SS, et al. The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol 2010;402:491-509. [Crossref] [PubMed]
- Wang WX, Danaher RJ, Miller CS, et al. Expression of miR-15/107 family microRNAs in human tissues and cultured rat brain cells. Genomics Proteomics Bioinformatics 2014;12:19-30. [Crossref] [PubMed]
- Bargaje R, Hariharan M, Scaria V, et al. Consensus miRNA expression profiles derived from interplatform normalization of microarray data. Rna 2010;16:16-25. [Crossref] [PubMed]
- Bruchova H, Yoon D, Agarwal AM, et al. Regulated expression of microRNAs in normal and polycythemia vera erythropoiesis. Exp Hematol 2007;35:1657-67. [Crossref] [PubMed]
- Choong ML, Yang HH, McNiece I. MicroRNA expression profiling during human cord blood-derived CD34 cell erythropoiesis. Exp Hematol 2007;35:551-64. [Crossref] [PubMed]
- Nelson PT, Baldwin DA, Kloosterman WP, et al. RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. Rna 2006;12:187-91. [Crossref] [PubMed]
- Wang Y, Luo J, Zhang H, et al. microRNAs in the Same Clusters Evolve to Coordinately Regulate Functionally Related Genes. Mol Biol Evol 2016;33:2232-47. [Crossref] [PubMed]
- Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A 2004;101:2999-3004. [Crossref] [PubMed]
- Shen X, Soibam B, Benham A, et al. miR-322/-503 cluster is expressed in the earliest cardiac progenitor cells and drives cardiomyocyte specification. Proc Natl Acad Sci U S A 2016;113:9551-6. [Crossref] [PubMed]
- Wilfred BR, Wang WX, Nelson PT. Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol Genet Metab 2007;91:209-17. [Crossref] [PubMed]
- Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 2014;42:D68-73. [Crossref] [PubMed]
- Nelson PT, Wang WX, Mao G, et al. Specific sequence determinants of miR-15/107 microRNA gene group targets. Nucleic Acids Res 2011;39:8163-72. [Crossref] [PubMed]
- Quann K, Jing Y, Rigoutsos I. Post-transcriptional regulation of BRCA1 through its coding sequence by the miR-15/107 group of miRNAs. Front Genet 2015;6:242. [Crossref] [PubMed]
- Cheng Y, Ji R, Yue J, et al. MicroRNAs are aberrantly expressed in hypertrophic heart: do they play a role in cardiac hypertrophy? Am J Pathol 2007;170:1831-40. [Crossref] [PubMed]
- Li F, Wang X, Capasso JM, et al. Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J Mol Cell Cardiol 1996;28:1737-46. [Crossref] [PubMed]
- Porrello ER, Johnson BA, Aurora AB, et al. MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ Res 2011;109:670-9. [Crossref] [PubMed]
- Nakamura M, Sadoshima J. Mechanisms of physiological and pathological cardiac hypertrophy. Nat Rev Cardiol 2018;15:387-407. [Crossref] [PubMed]
- van Rooij E, Sutherland LB, Liu N, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A 2006;103:18255-60. [Crossref] [PubMed]
- You XY, Huang JH, Liu B, et al. HMGA1 is a new target of miR-195 involving isoprenaline-induced cardiomyocyte hypertrophy. Biochemistry (Mosc) 2014;79:538-44. [Crossref] [PubMed]
- Zhang X, Liu Y, Han Q. Puerarin Attenuates Cardiac Hypertrophy Partly Through Increasing Mir-15b/195 Expression and Suppressing Non-Canonical Transforming Growth Factor Beta (Tgfβ) Signal Pathway. Med Sci Monit 2016;22:1516-23. [Crossref] [PubMed]
- Wang L, Qin D, Shi H, et al. MiR-195-5p Promotes Cardiomyocyte Hypertrophy by Targeting MFN2 and FBXW7. Biomed Res Int 2019;2019:1580982. [Crossref] [PubMed]
- Zhang X, Ji R, Liao X, et al. MicroRNA-195 Regulates Metabolism in Failing Myocardium Via Alterations in Sirtuin 3 Expression and Mitochondrial Protein Acetylation. Circulation 2018;137:2052-67. [Crossref] [PubMed]
- Busk PK, Cirera S. MicroRNA profiling in early hypertrophic growth of the left ventricle in rats. Biochem Biophys Res Commun 2010;396:989-93. [Crossref] [PubMed]
- Ishihara N, Eura Y, Mihara K. Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. J Cell Sci 2004;117:6535-46. [Crossref] [PubMed]
- Guan X, Wang L, Liu Z, et al. miR-106a promotes cardiac hypertrophy by targeting mitofusin 2. J Mol Cell Cardiol 2016;99:207-17. [Crossref] [PubMed]
- Lv L, Li T, Li X, et al. The lncRNA Plscr4 Controls Cardiac Hypertrophy by Regulating miR-214. Mol Ther Nucleic Acids 2018;10:387-97. [Crossref] [PubMed]
- Peoples JN, Saraf A, Ghazal N, et al. Mitochondrial dysfunction and oxidative stress in heart disease. Exp Mol Med 2019;51:1-13. [Crossref] [PubMed]
- Chen H, Untiveros GM, McKee LA, et al. Micro-RNA-195 and -451 regulate the LKB1/AMPK signaling axis by targeting MO25. PLoS One 2012;7:e41574. [Crossref] [PubMed]
- Thygesen K, Alpert JS, Jaffe AS, et al. Third universal definition of myocardial infarction. J Am Coll Cardiol 2012;60:1581-98. [Crossref] [PubMed]
- Burgos JI, Morell M, Mariángelo JIE, et al. Hyperosmotic stress promotes endoplasmic reticulum stress-dependent apoptosis in adult rat cardiac myocytes. Apoptosis 2019;24:785-97. [Crossref] [PubMed]
- Gao CK, Liu H, Cui CJ, et al. Roles of MicroRNA-195 in cardiomyocyte apoptosis induced by myocardial ischemia-reperfusion injury. J Genet 2016;95:99-108. [Crossref] [PubMed]
- Hang P, Sun C, Guo J, et al. BDNF-mediates Down-regulation of MicroRNA-195 Inhibits Ischemic Cardiac Apoptosis in Rats. Int J Biol Sci 2016;12:979-89. [Crossref] [PubMed]
- Chen C, Jia KY, Zhang HL, et al. MiR-195 enhances cardiomyocyte apoptosis induced by hypoxia/reoxygenation injury via downregulating c-myb. Eur Rev Med Pharmacol Sci 2016;20:3410-6. [PubMed]
- Zhang N, Meng X, Mei L, et al. The Long Non-Coding RNA SNHG1 Attenuates Cell Apoptosis by Regulating miR-195 and BCL2-Like Protein 2 in Human Cardiomyocytes. Cell Physiol Biochem 2018;50:1029-40. [Crossref] [PubMed]
- Zhu H, Yang Y, Wang Y, et al. MicroRNA-195 promotes palmitate-induced apoptosis in cardiomyocytes by down-regulating Sirt1. Cardiovasc Res 2011;92:75-84. [Crossref] [PubMed]
- Porrello ER, Mahmoud AI, Simpson E, et al. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc Natl Acad Sci U S A 2013;110:187-92. [Crossref] [PubMed]
- Miki T, Yuda S, Kouzu H, et al. Diabetic cardiomyopathy: pathophysiology and clinical features. Heart Fail Rev 2013;18:149-66. [Crossref] [PubMed]
- Zheng D, Ma J, Yu Y, et al. Silencing of miR-195 reduces diabetic cardiomyopathy in C57BL/6 mice. Diabetologia 2015;58:1949-58. [Crossref] [PubMed]
- Khakdan S, Delfan M, Heydarpour Meymeh M, et al. High-intensity interval training (HIIT) effectively enhances heart function via miR-195 dependent cardiomyopathy reduction in high-fat high-fructose diet-induced diabetic rats. Arch Physiol Biochem 2020;126:250-7. [Crossref] [PubMed]
- Shah AM, Mann DL. In search of new therapeutic targets and strategies for heart failure: recent advances in basic science. Lancet 2011;378:704-12. [Crossref] [PubMed]
- Hill JA, Olson EN. Cardiac plasticity. N Engl J Med 2008;358:1370-80. [Crossref] [PubMed]
- Shen Y, Zhang W, Lee L, et al. Down-regulated microRNA-195-5p and up-regulated CXCR4 attenuates the heart function injury of heart failure mice via inactivating JAK/STAT pathway. Int Immunopharmacol 2020;82:106225. [Crossref] [PubMed]
- Xie C, Qi H, Huan L, et al. Effect of miR-195-5p on cardiomyocyte apoptosis in rats with heart failure by regulating TGF-β1/Smad3 signaling pathway. Biosci Rep 2020;40:BSR20200566. [Crossref] [PubMed]
- Xing Y, Liu Z, Yang G, et al. MicroRNA expression profiles in rats with selenium deficiency and the possible role of the Wnt/β-catenin signaling pathway in cardiac dysfunction. Int J Mol Med 2015;35:143-52. [Crossref] [PubMed]
- Jafarzadeh M, Mohammad Soltani B, Ekhteraei Tousi S, et al. Hsa-miR-497 as a new regulator in TGFβ signaling pathway and cardiac differentiation process. Gene 2018;675:150-6. [Crossref] [PubMed]
- Xiao Y, Zhang X, Fan S, et al. MicroRNA-497 Inhibits Cardiac Hypertrophy by Targeting Sirt4. PLoS One 2016;11:e0168078. [Crossref] [PubMed]
- Zhu Y, Yan Y, Principe DR, et al. SIRT3 and SIRT4 are mitochondrial tumor suppressor proteins that connect mitochondrial metabolism and carcinogenesis. Cancer Metab 2014;2:15. [Crossref] [PubMed]
- Verdin E, Hirschey MD, Finley LW, et al. Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends Biochem Sci 2010;35:669-75. [Crossref] [PubMed]
- Li X, Zeng Z, Li Q, et al. Inhibition of microRNA-497 ameliorates anoxia/reoxygenation injury in cardiomyocytes by suppressing cell apoptosis and enhancing autophagy. Oncotarget 2015;6:18829-44. [Crossref] [PubMed]
- Qin L, Yang W, Wang YX, et al. MicroRNA-497 promotes proliferation and inhibits apoptosis of cardiomyocytes through the downregulation of Mfn2 in a mouse model of myocardial ischemia-reperfusion injury. Biomed Pharmacother 2018;105:103-14. [Crossref] [PubMed]
- Ghosh G, Subramanian IV, Adhikari N, et al. Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-α isoforms and promotes angiogenesis. J Clin Invest 2010;120:4141-54. [Crossref] [PubMed]
- Tsai SH, Huang PH, Tsai HY, et al. Roles of the hypoximir microRNA-424/322 in acute hypoxia and hypoxia-induced pulmonary vascular leakage. Faseb j 2019;33:12565-75. [Crossref] [PubMed]
- Lou Y, Wang S, Qu J, et al. miR-424 promotes cardiac ischemia/reperfusion injury by direct targeting of CRISPLD2 and regulating cardiomyocyte pyroptosis. Int J Clin Exp Pathol 2018;11:3222-35. [PubMed]
- Huang S, Zou X, Zhu JN, et al. Attenuation of microRNA-16 derepresses the cyclins D1, D2 and E1 to provoke cardiomyocyte hypertrophy. J Cell Mol Med 2015;19:608-19. [Crossref] [PubMed]
- Wang X, Shang Y, Dai S, et al. MicroRNA-16-5p Aggravates Myocardial Infarction Injury by Targeting the Expression of Insulin Receptor Substrates 1 and Mediating Myocardial Apoptosis and Angiogenesis. Curr Neurovasc Res 2020;17:11-7. [Crossref] [PubMed]
- Liu J, Sun F, Wang Y, et al. Suppression of microRNA-16 protects against acute myocardial infarction by reversing beta2-adrenergic receptor down-regulation in rats. Oncotarget 2017;8:20122-32. [Crossref] [PubMed]
- Keeley EC, Mehrad B, Janardhanan R, et al. Elevated circulating fibrocyte levels in patients with hypertensive heart disease. J Hypertens 2012;30:1856-61. [Crossref] [PubMed]
- Dickinson BA, Semus HM, Montgomery RL, et al. Plasma microRNAs serve as biomarkers of therapeutic efficacy and disease progression in hypertension-induced heart failure. Eur J Heart Fail 2013;15:650-9. [Crossref] [PubMed]
- He L, Huang C. MiR-19b and miR-16 cooperatively signaling target the regulator ADRA1A in Hypertensive heart disease. Biomed Pharmacother 2017;91:1178-83. [Crossref] [PubMed]
- Liu LF, Liang Z, Lv ZR, et al. MicroRNA-15a/b are up-regulated in response to myocardial ischemia/reperfusion injury. J Geriatr Cardiol 2012;9:28-32. [Crossref] [PubMed]
- Roy S, Khanna S, Hussain SR, et al. MicroRNA expression in response to murine myocardial infarction: miR-21 regulates fibroblast metalloprotease-2 via phosphatase and tensin homologue. Cardiovasc Res 2009;82:21-9. [Crossref] [PubMed]
- van Rooij E, Sutherland LB, Thatcher JE, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A 2008;105:13027-32. [Crossref] [PubMed]
- Hullinger TG, Montgomery RL, Seto AG, et al. Inhibition of miR-15 protects against cardiac ischemic injury. Circ Res 2012;110:71-81. [Crossref] [PubMed]
- Liu L, Zhang G, Liang Z, et al. MicroRNA-15b enhances hypoxia/reoxygenation-induced apoptosis of cardiomyocytes via a mitochondrial apoptotic pathway. Apoptosis 2014;19:19-29. [Crossref] [PubMed]
- Nishi H, Ono K, Iwanaga Y, et al. MicroRNA-15b modulates cellular ATP levels and degenerates mitochondria via Arl2 in neonatal rat cardiac myocytes. J Biol Chem 2010;285:4920-30. [Crossref] [PubMed]
- Taegtmeyer H, Razeghi P, Young ME. Mitochondrial proteins in hypertrophy and atrophy: a transcript analysis in rat heart. Clin Exp Pharmacol Physiol 2002;29:346-50. [Crossref] [PubMed]
- Aoyama T, Matsui T, Novikov M, et al. Serum and glucocorticoid-responsive kinase-1 regulates cardiomyocyte survival and hypertrophic response. Circulation 2005;111:1652-9. [Crossref] [PubMed]
- Tijsen AJ, van der Made I, van den Hoogenhof MM, et al. The microRNA-15 family inhibits the TGFβ-pathway in the heart. Cardiovasc Res 2014;104:61-71. [Crossref] [PubMed]
- Liu Z, Yang D, Xie P, et al. MiR-106b and MiR-15b modulate apoptosis and angiogenesis in myocardial infarction. Cell Physiol Biochem 2012;29:851-62. [Crossref] [PubMed]
- Qi H, Ren J, E M, et al. MiR-103 inhibiting cardiac hypertrophy through inactivation of myocardial cell autophagy via targeting TRPV3 channel in rat hearts. J Cell Mol Med 2019;23:1926-39. [Crossref] [PubMed]
- Rech M, Kuhn AR, Lumens J, et al. AntagomiR-103 and -107 Treatment Affects Cardiac Function and Metabolism. Mol Ther Nucleic Acids 2019;14:424-37. [Crossref] [PubMed]
- Trajkovski M, Hausser J, Soutschek J, et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 2011;474:649-53. [Crossref] [PubMed]
- Wang JX, Zhang XJ, Li Q, et al. MicroRNA-103/107 Regulate Programmed Necrosis and Myocardial Ischemia/Reperfusion Injury Through Targeting FADD. Circ Res 2015;117:352-63. [Crossref] [PubMed]
- Sayed D, Hong C, Chen IY, et al. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res 2007;100:416-24. [Crossref] [PubMed]
- Gupta SK, Bang C, Thum T. Circulating microRNAs as biomarkers and potential paracrine mediators of cardiovascular disease. Circ Cardiovasc Genet 2010;3:484-8. [Crossref] [PubMed]
- Jin P, Gu W, Lai Y, et al. The Circulating MicroRNA-206 Level Predicts the Severity of Pulmonary Hypertension in Patients with Left Heart Diseases. Cell Physiol Biochem 2017;41:2150-60. [Crossref] [PubMed]
- Verma AM, Patel M, Aslam MI, et al. Circulating plasma microRNAs as a screening method for detection of colorectal adenomas. Lancet 2015;385 Suppl 1:S100. [Crossref] [PubMed]
- Abbas NA, John RI, Webb MC, et al. Cardiac troponins and renal function in nondialysis patients with chronic kidney disease. Clin Chem 2005;51:2059-66. [Crossref] [PubMed]
- Long G, Wang F, Duan Q, et al. Circulating miR-30a, miR-195 and let-7b associated with acute myocardial infarction. PLoS One 2012;7:e50926. [Crossref] [PubMed]
- Jaguszewski M, Osipova J, Ghadri JR, et al. A signature of circulating microRNAs differentiates takotsubo cardiomyopathy from acute myocardial infarction. Eur Heart J 2014;35:999-1006. [Crossref] [PubMed]
- Mazzone AL, Baker RA, McNicholas K, et al. Circulating and Urinary miR-210 and miR-16 Increase during Cardiac Surgery Using Cardiopulmonary Bypass - A Pilot Study. J Extra Corpor Technol 2018;50:19-29. [PubMed]
- Ellis KL, Cameron VA, Troughton RW, et al. Circulating microRNAs as candidate markers to distinguish heart failure in breathless patients. Eur J Heart Fail 2013;15:1138-47. [Crossref] [PubMed]
- Vegter EL, Schmitter D, Hagemeijer Y, et al. Use of biomarkers to establish potential role and function of circulating microRNAs in acute heart failure. Int J Cardiol 2016;224:231-9. [Crossref] [PubMed]
- Voellenkle C, van Rooij J, Cappuzzello C, et al. MicroRNA signatures in peripheral blood mononuclear cells of chronic heart failure patients. Physiol Genomics 2010;42:420-6. [Crossref] [PubMed]
- Adhikari N, Guan W, Capaldo B, et al. Identification of a new target of miR-16, Vacuolar Protein Sorting 4a. PLoS One 2014;9:e101509. [Crossref] [PubMed]
- Marques FZ, Vizi D, Khammy O, et al. The transcardiac gradient of cardio-microRNAs in the failing heart. Eur J Heart Fail 2016;18:1000-8. [Crossref] [PubMed]
- Fang L, Ellims AH, Moore XL, et al. Circulating microRNAs as biomarkers for diffuse myocardial fibrosis in patients with hypertrophic cardiomyopathy. J Transl Med 2015;13:314. [Crossref] [PubMed]
- Wu J, Li J, Liu H, et al. Circulating plasma circular RNAs as novel diagnostic biomarkers for congenital heart disease in children. J Clin Lab Anal 2019;33:e22998. [Crossref] [PubMed]
- Janssen HL, Reesink HW, Lawitz EJ, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med 2013;368:1685-94. [Crossref] [PubMed]
- Jopling CL, Yi M, Lancaster AM, et al. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 2005;309:1577-81. [Crossref] [PubMed]
- Montgomery RL, Hullinger TG, Semus HM, et al. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation 2011;124:1537-47. [Crossref] [PubMed]
- Acunzo M, Croce CM. Downregulation of miR-15a and miR-16-1 at 13q14 in Chronic Lymphocytic Leukemia. Clin Chem 2016;62:655-6. [Crossref] [PubMed]
- Luo C, Tetteh PW, Merz PR, et al. miR-137 inhibits the invasion of melanoma cells through downregulation of multiple oncogenic target genes. J Invest Dermatol 2013;133:768-75. [Crossref] [PubMed]