
Long non-coding RNAs: novel players in the pathogenesis of polycystic ovary syndrome
Introduction
Polycystic ovary syndrome (PCOS), a World Health Organization group II ovulation disorder, is one of the most common endocrinological disorders in reproductive-aged women with a prevalence ranging between 8% and 13% (1,2). Its cardinal features are oligo- or anovulation, clinical and/or biochemical signs of hyperandrogenism, and polycystic ovaries (3). In addition, women with PCOS may present with other disorders, including insulin resistance (IR), obesity, type 2 diabetes, cardiovascular disease and anxiety (1-3). Given the diversities and complications of this condition, a broader picture of its etiology needs to be elucidated.
More recently, the understanding of PCOS has been enhanced by genome and transcriptomic analyses in PCOS patients with the potential to identify the etiological factors underpinning this disease and opening precision medicine approaches to its management. However, it is still a challenge to characterize the full processes of multiple molecular interactions on gene regulatory networks in PCOS, specifically in understanding the spectrum of DNA, RNA and protein interactions.
We hypothesized that long noncoding RNAs (lncRNAs) might play an important role in the pathogenesis of PCOS. LncRNAs are defined as RNA transcripts of length >200 nucleotides that lack protein-coding capacity, and about 60,000 lncRNAs are contained in human genome (4,5). LncRNAs have been characterized as functional RNA, involved in biological, developmental and pathological processes, acting through mechanisms such as epigenetics, cis regulation at enhancers and post-transcriptional regulation of mRNA processing (5). More recently, studies concerned with the functions of lncRNAs in PCOS have suggested new approaches to understanding the pathogenesis of this condition.
In this review, we discuss the dysregulation of lncRNAs in PCOS and the potential influence of lncRNAs on current and future diagnosis and therapies.
LncRNAs structure and functions
With the emergence of high-throughput sequencing, many lncRNAs have been identified. Like other non-coding RNAs (ncRNAs), their unique but complex three-dimensional structure enables lncRNAs to exhibit a wide-range of biological functions.
Biogenesis and structure
Based on their size, non-coding RNAs can be divided into small non-coding RNAs and long non-coding RNAs (>200 nucleotides). LncRNAs are generated from 80–90% non-coding regulatory elements in the human genome (6-8). Other ncRNAs include transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), small interfering RNAs (siRNAs), microRNAs (miRNAs), piwiinteracting RNAs (piRNAs), small nucleolar RNAs (snoRNAs) and small nuclear RNAs (snRNAs).
LncRNAs share similar biogenesis pathways as other ncRNAs and are generated by RNA polymerase II, polyadenylated, and spliced. Owing to their positional relationship to protein-coding genes, lncRNAs can be flexibly transcribed from different genomic regions, i.e., promoter upstream, enhancers, intergenic, and the opposite strand of protein-coding genes (9). In this manner, lncRNAs are classified into five groups (10): (I) stand-alone lncRNAs; (II) natural anti-sense transcripts; (III) pseudogenes; (IV) intronic transcript; (V) divergent transcripts, promoter-associated transcripts, and enhancer RNAs. In addition, many lncRNAs are also generated from long primary transcripts with unusual RNA processing, yielding distinctive structures. For example, instead of canonical RNA maturation processing, lncRNAs could generate a complete 3' end by ribonuclease P cleavage to generate the stable format (11), capped by small nucleolar RNA (snoRNA)-protein complexes at their ends (12), or generate isoforms through alternative pathways (13). By comparison to mRNA, lncRNAs lack open reading frames (ORFs) but have special 3-terminal processes and a greater tendency towards cell type-specific expression (14,15). Finally, both are degraded by exonuclease degradation or exosome digestion (15).
Due to the WatsonCrick base pairing and base stacking, lncRNAs transcripts can fold into highly stable secondary and tertiary structures (14). RNA has the capacity to form double-stranded stems, single-stranded loops and bulges, which energetically fold further into high-order and complex 3D architectures (9,16). These distinctive structures enable lncRNAs to be identified amongst mRNAs or other ncRNAs allowing their interaction with DNA or other RNA molecules, even forming the lncRNA-protein complexes. A prominent example is the hox transcript antisense RNA (HOTAIR), originating from the HOXC locus, which can serve as scaffolds by combining with related histone modification enzymes (i.e., the 5' domain binds PRC2 while the 3' domain binds the LSD1), to assist post-transcriptional modifications (17), suggesting lncRNAs as essential molecular players in diverse human physiological and pathological processes.
Main biological functions
Since the first functional lncRNAs were discovered in the 1990s (i.e., H19, Xist) (10,18), substantial research has delineated abundant structurefunction relationships of lncRNAs, and the concept of “transcriptional noise” (the old argument about whether ncRNAs have functions) in this field (19). However, the complexity and diversity of lncRNAs three-dimensional structure, as well as their expression specificity in different cell types and developmental stages (20,21), have resulted in the difficulty of categorizing the myriad functions of lncRNAs. Nevertheless, according to existing functional studies, the molecular mechanisms of lncRNAs are divided into four types: signal, decoy, guide, and scaffold (22). A more common understanding about the role of lncRNAs in regulatory mechanisms is at the level of chromatin modification, transcription and post-transcriptional processing (23).
The role of lncRNAs in the epigenetic regulation has been of considerable interest. Many lncRNAs regulate the chromatin states via recruiting chromatin-modifying enzyme proteins to specific sites in the gene (24). LncRNAs can act on their target genes through cis-regulation, affecting expression of the neighboring gene on the same allele from which they are transcribed. In trans-regulation, lncRNAs do not meet this criterion (25). For instance, lncRNA Xist accumulates in cis on the X chromosome and recruits PRC2, provoking the stable silencing of the entire chromosome (26), while HOTAIR interacts with PRC2 to repress transcription of the HOXD locus in trans (27). Additionally, lncRNAs acting as scaffolds can assemble multiple proteins to form lncRNA - ribonucleoprotein complexes, affecting chromatin modifications, such as HOTAIR.
Given that lncRNAs can be generated from promoters or enhancers, transcription can be affected, both positively and negatively. Those lncRNAs can act as decoys for transcription factors. LncRNAs can act as coregulators modulating transcription factor activity. LncRNA steroid receptor RNA activator (SRA) is considered as a coactivator for a number of nuclear steroid receptors (28). Some lncRNAs interface with additional regulators (i.e., ATP-dependent chromatin remodeling complexes and histone acetyltransferases) to modulate gene expression (29). In addition, lncRNAs can directly interfere with Pol II activity by combining with the initiation complex to influence promoter choice (30).
As functional RNA, lncRNAs play an essential role in post-transcriptional processing, including splicing, editing, translation and degradation. The lncRNA MALAT1, highly enriched in nuclear speckles, has been suggested to be a general component of the splicing machinery and regulates the phosphorylation level of serine/arginine (SR) splicing factors, thus, controlling alternative splicing of certain mRNA precursors (31,32). Moreover, circRNAs, a special class of lncRNAs, defined by their circular structure, are resistant to degradation by exonucleases, and are highly stable within a cell (33).
Taken together, the role of lncRNAs is extremely broad across the biological processes. The greater understanding of lncRNA structure and function is still evolving. However, the development of pioneering sequencing technologies could depict more secondary structures, proteinbinding motifs and other features about lncRNAs, which may reveal additional cellular functions.
LncRNAs in physiology of female fecundity
Recently short ncRNAs have been shown to play an important role in the control of reproductive functions (34,35). However, only limited reports exist on the potential regulatory impact of lncRNA in female fecundity. LncRNA nuclear paraspeckle assembly transcript 1 (NEAT1) is an essential component of paraspeckle, participating in the regulation of transcription and RNA processing (36-38). In support of the physiological function of lncRNA and paraspeckle, Neat1 knockout (KO) mice failed to become pregnant despite normal ovulation. However, they expressed corpus luteum dysfunction and with low progesterone values (38). The H19 KO mice demonstrated subfertility and accelerated follicular recruitment, implying that H19 limited the number of follicles that mature, produce estradiol, and ovulate (39).
The developmental capacity of mature oocytes is essential for maintaining the reproductive potential of the female, resulting from multiple molecular interactions in the follicular microenvironment [i.e., the oocyte, the surrounding follicular cells and follicular fluid (FF)] (40,41). Differential lncRNA expression profiles in human cumulus-oocyte complex (COC) were recently analyzed. These studies found abundant lncRNA expression in human MII oocytes (BCAR4, C3orf56, TUNAR, OOEP-AS1, CASC18, and LINC01118) and cumulus cells (CCs) (NEAT1, MALAT1, ANXA2P2, MEG3, IL6STP1, and VIM-AS1) (42). An independent microarray analysis was performed on FF and revealed that four lncRNAs (RP1-34H18.1, LNC-000503, LNC-000683, H19) differed between mature and immature ovarian follicles of healthy women (43). The interrogation of transcriptome dynamics in human primordial, primary and small antral follicles uncovered the expression of lncRNAs XIST, NEAT1, NEAT2 (MALAT1), and GAS5 (44). These lncRNAs are indispensable during development of the normal human follicle but the functions remain to be determined. An additional important lncRNA in human oocyte is telomeric repeat containing RNA (TERRA), an essential component of telomeric architecture, colocalizing with telomeres and telomerase during meiotic prophase I (45). Both TERRA levels and distribution would be regulated in a gender-specific manner (45). TERRA levels were markedly lower in human fetal oocytes compared to spermatocytes (45,46). The authors speculated that TERRA could contribute to telomeric stability during oogenesis and may be related to telomerase function during follicular development (46).
These studies suggest that several distinct lncRNAs are present during human follicular development but their detailed role remains to be determined
LncRNAs in women with PCOS
LncRNAs as a novel class of molecules have attracted attention in the pathophysiology of human diseases, and multiple functional studies of lncRNAs have focused on tumor pathophysiology. Recently, different lncRNAs have been detected in PCOS, and many studies have described numerous lncRNAs in various biological and pathological processes of granulosa cells (GCs), FF, peripheral blood and ovaries (Table 1; Figure 1).
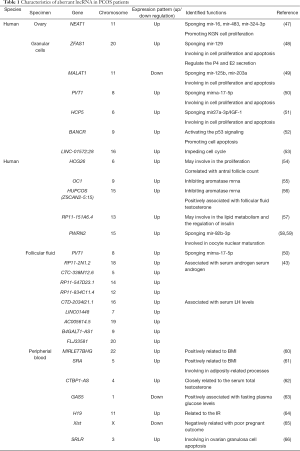
Full table
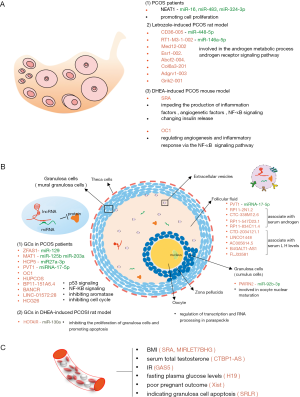
LncRNAs in human GCs
GCs, one of somatic cells in follicles, are divided into mural granular cells (MGCs, line the wall of follicle) and CCs (directly surround the oocyte) until antrum formation (67). GCs are the most studied cell type in PCOS. Usually, GCs are collected from women with or without PCOS undergoing in vitro fertilization (IVF) or intracytoplasmic sperm injection (ICSI) to determine the potentially pathogenic genes. LncRNAs have been involved in diverse manifestations of PCOS mainly through the interaction between lncRNA and miRNA (Figure 1).
Cell proliferation and/or apoptosis
Currently, several lncRNAs have been found to play a vital molecular role in regulating GC proliferation and apoptosis by functioning as the competitive endogenous RNA (ceRNA). The level of lncRNA zinc finger antisense 1 (ZFAS1), located on chromosome 20q13, is elevated in ovarian GCs of PCOS patients. LncRNA ZFAS1 combining with miR-129 prevents miRNA-mediated degradation of HMGB1, inhibiting the proliferation activity while promoting the apoptosis of GCs (48). The expression of NEAT1 is upregulated in PCOS ovarian tissue, promoting the proliferation of human granulosa-like tumor cell line KGN via several miRNAs (i.e., miR-16, miR-483 and miR-324-3p) (47). However, whether the upregulated NEAT1 causes corpus luteum dysfunction in PCOS, like NEAT1 KO mice (38), is unclear. Decreased lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) in women with PCOS, as a downstream transcript of NEAT1, destroys the cell cycle in GCs by regulating the TGF-βsignaling pathway via ceRNA network (49). Interestingly, MALAT1 KO female mice have no any obvious abnormal reproductive phenotype (68). Whether the down-regulated MALTA1 exerts separate functions in different species remains to be determined. Elevated lncRNA PVT1, located in the nuclei, regulates the expression of PTEN by interacting with miRNA-17-5p in the apoptosis and proliferation processing of GCs (50). Increased lncRNA HCP5 also is involved in the GC proliferation and apoptosis via sponging miR27a-3p/IGF-1 (51). In addition, several other signal pathways have been reported to be involved in the regulation of GC proliferation and apoptosis. LncRNA BANCR promotes GC apoptosis in PCOS by activating the p53 signaling pathway (52). LINC-01572:28 hinders the interaction between SKP2 and P27 to suppress GC proliferation (53). LncRNA HCG26 is found by microarray and possibly contributes to GC proliferation, but the specific molecular mechanism of this lncRNA is not known (54).
Oocyte development
Huang et al. firstly described the dysregulated lncRNAs in CCs of women with PCOS via microarray analysis, and they found the differentially expressed lncRNAs with the length between 201 to 11,869 bp were mostly transcribed from chromosome 2. Co-expression network analysis of 43 lncRNAs and 29 mRNAs found that aberrantly expressed lncRNAs may be involved in the regulation of oocyte development (58). Up-regulated lncRNA Prader-Willi region nonprotein coding RNA 2 (PWRN2) in CCs is involved in oocyte nuclear maturation through the PWRN2/miR-92b-3p/TMEM120B ceRNA network (59). Elevated lncRNA HCG26 is correlated with antral follicle count (AFC) inspected by ultrasonic B, suggesting that dysregulated lncRNA may contribute to follicle development (54). However, understanding the specific role of lncRNAs in the molecular mechanism of oocyte maturation requires further elucidation.
Steroidogenesis
PCOS is such a heterogeneous disorder that there are various clinical manifestations, including hyperandrogenemia, obesity and IR. LncRNAs are involved in the regulation of steroidogenesis and metabolism. Downregulated lncRNA ZFAS1 is reported to increase P4 and E2 secretion in GCs (48). The knockdown of lncRNA OC1 identified in human GCs could increase the expression of aromatase mRNA, leading to increased production of estradiol (55). LncRNA HUPCOS, located in 15q22, is a new identified intergenic lncRNA with a 495bp transcript length in GCs. LncRNA HUPCOS is reported to be positively associated with FF testosterone through inhibition of CYP11 activity (56).
Relationship with PCOS clinical phenotypes
An increase of lncRNA RP11-151A6.4 expression is observed in GCs from PCOS patients with higher BMI, higher HOMA-IR values and hyperinsulinemia, indicating that the intronic lncRNA RP11-151A6.4 may be involved in lipid metabolism and the regulation of insulin (57). Differentially expressed lncRNAs in GCs isolated from the PCOS patients with or without hyperandrogenism may play an important role in metabolism via regulating mitochondrial function (69).
These insights indicate that lncRNAs may play a key role in GCs through different signal pathways, and drive the ovulation disorder and emergence of infertility in PCOS. Therefore, lncRNAs could be a potential therapeutic target.
LncRNAs in human FF
The development of the antrum of follicle requires the influx of water, which gradually forms the FF. The microenvironment of FF is vital to nourish oocyte growth and follicular maturation. The FF contains miRNA, proteins, various ionic compounds and metabolic products (70). Recently, a high-throughput RNA-seq assay in human FF of mature or immature follicles spots 1,583 novel lncRNAs that are enriched in metabolism and cell-cell connection related processes. Among the differentially expressed lncRNAs, four kinds of lncRNAs are associated with serum androgen and another five kinds of lncRNA are associated with serum LH levels (43). LncRNA PVT1 is increased in FF, but the mechanism is not known (50). Extracellular vesicles (EVs) have been reported to transport miRNAs and proteins to recipient cells in the ovarian follicle (71), but without lncRNAs. We assume that the lncRNAs in FF are possibly derived from EVs and regulate gene expression in GCs or oocytes during the follicle development.
LncRNAs in human peripheral blood
The blood circulation is the transportation hub for different tissue or cell metabolites in humans. LncRNAs in peripheral blood can partially reflect the expression of lncRNAs in vivo. Therefore, the abnormal expression of lncRNA in periphery blood is expected to serve as a biomarker for the diagnosis of PCOS.
Several lncRNAs in peripherial blood vary with metabolic phenotypes in PCOS, such as obesity, hyperandrogenism and IR. Both lncRNA SRA and MIRLET7BHG are observed to be elevated in peripheral blood of women with PCOS and are positively related to BMI (60,61). Furthermore, lncRNA SRA is thought to be an important mediator in adiposity-related processes in PCOS (61). LncRNA terminal binding protein 1 antisense (CTBP1-AS) serves as a novel androgen receptor modulator and is closely related to the serum total testosterone (62). LncRNA growth-arrest specific transcript 5 (GAS5) is detected with decreased expression in the serum of PCOS patients with IR, whilst interleukin-18 (IL-18) is significantly increased, indicating that IL-18 and GAS5 may contribute to IR in PCOS (63). Another lncRNA H19 is positively associated with fasting plasma glucose levels, but has no correlation with IR (64). Evidence has accumulated showing a vital link between lncRNAs and pregnancy outcomes. For instance, decreased lncRNA X-inactive specific transcript (Xist) is significantly related with adverse pregnancy outcome in PCOS patients, including gestational diabetes, pre-eclampsia, caesarean section, preterm and post-term birth (65). In addition, some up-regulated lncRNAs in blood may reflect the ovarian GC apoptosis, such as lncRNA sorafenib resistance-associated lncRNA in renal cell carcinoma (SRLR) (66) may reflect ovarian GC apoptosis.
Taken together, these studies provide new insights for our further understandings of lncRNAs in PCOS and expand the novel diagnostic biomarkers and therapeutic targets.
LncRNAs in PCOS rodent models
Various animal models in rats or mice have been established to examine the potential physiopathologic mechanism in PCOS. The main protocols induce the hyperandrogenism and metabolic abnormalities (72). Accordingly, we outline the lncRNA studies in three different PCOS rodent models that are induced by three methods, including dehydroepiandrosterone (DHEA) and letrozole (Table 2; Figure 1).
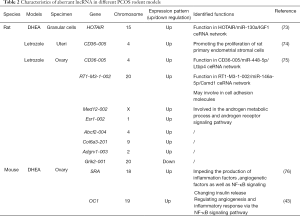
Full table
LncRNAs in DHEA-induced rodent models
PCOS rats or mice with subcutaneous DHEA for 20 consecutive days (73,76). In the DHEA-induced rat model, serum levels of estradiol (E2), testosterone (T), luteinizing hormone (LH) and follicle-stimulating hormone (FSH) were similar to PCOS patients. LncRNA HOTAIR increased the expression of IGF1 in GCs via competitive binding to miR-130a, inhibiting their proliferation and promoting apoptosis (73). In the DHEA-induced PCOS mice, lncRNA SRA knockdown in the ovary impedes the production of inflammation factors, angiogenetic factors and the NF-κB signaling pathway and changes insulin release (76). LncRNA OC1, up-regulated in human GCs, also increases in PCOS mice ovaries and positively regulates serum insulin release and production of angiogenesis-related factors through NF-κB signaling pathway (55).
LncRNAs in letrozole-induced rodent models
Eighty-seven-day-old SD rats were administered with letrozole for 23 consecutive days (75). This PCOS model is consistent with phenotypes of the PCOS patient, including obesity, irregular estrous cycle, higher androgen and LH in serum and more cystic follicles in the ovaries. A total of 158 lncRNAs were differently expressed in the ovary tissue of PCOS rats via RNA sequencing analysis. Up-regulated lncRNA Med12-002 and lncRNA Esr1-002 are involved in the androgen metabolic process and androgen receptor signaling pathway. LncRNA RT1-M3-1-002/miR-146a-5p/Csmd1 and lncRNA CD36-005/miR-448-5p/Ltbp4 are present in the PCOS mice (75). In the same PCOS rat model, lncRNA CD36-005 is up-regulated in the uterus and promotes the proliferation of rat primary endometrial stromal cells (74).
Consequently, lncRNAs are involved in multiple metabolic mechanisms in PCOS rodent models. However, it is yet unclear whether expression of specific lncRNAs is distinctly associated with PCOS phenotypes. To date, the literature about lncRNAs in PCOS rodent models is still rare. Thus, there are a glut of pathophysiology to be uncovered.
Conclusions and perspectives
LncRNAs are new players that are shaping the molecular configuration in pathogenesis of PCOS. Due to the manifold and complex three-dimensional structure, lncRNAs are playing an indispensable role in amounts of biological process, enhancing the understanding of pathological mechanism in PCOS. However, available information about lncRNA molecular mechanisms is still fragmented in PCOS. Compared with their roles in physiology, the dynamic expression process of preponderant lncRNAs at different stages of follicle is unclear in PCOS. Also, there is insufficient evidence to suggest that an abnormal lncRNA can be correlated one-to-one with a clinical phenotype. Based on gene therapy, we should classify aberrant lncRNAs in different phenotypes of PCOS and establish a database for the precision management in clinical practice.
Overall, the investigations about lncRNAs in PCOS provided a new vision for the comprehension of the pathophysiology mechanism. In the future, lncRNAs should be examined as circulating biomarkers for detection of PCOS and precise treatment.
Limitations
Articles about PCOS quoted in this review are all from the Chinese literature and there is a bias in population distribution. Considering the diversity of clinical manifestations of PCOS, differentially expressed lncRNA should be more precisely analyzed for disease subtypes. Increasing evidence supports the role of lncRNAs in normal follicle development. Nevertheless, our knowledge of lncRNAs in oocytes of women with PCOS is almost non-existent, mainly due to the difficulty in obtaining clinical specimens. Further, the dysregulated lncRNAs in PCOS animal models are not necessarily suitable for all mechanisms of PCOS clinical phenotypes. Apart from the ceRNA network, lncRNA can also function in epigenetic regulation and alter the transcription or post-transcriptional landscape. It is clearly necessary to study the function of lncRNA from more perspectives in PCOS.
Acknowledgments
We are deeply grateful to professor John R. G. Challis (University of Toronto), professor Yiping Shen (Harvard Medical School) and our colleague Siwen Wang (Harvard Medical School) for their editing of the manuscript.
Funding: This study was supported by grants from the National Key Research & Developmental Program of China (No. 2017 YFC 1001003), National Natural Science Foundation of China (No. 81471421).
Footnote
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-5044
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at: http://dx.doi.org/10.21037/atm-20-5044). Dr. DZ, Dr. LM, Dr. YW, and MT report grants from National Key Research & Developmental Program of China, grants from National Natural Science Foundation of China, during the conduct of the study.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Teede HJ, Misso ML, Costello MF, et al. Recommendations from the international evidence-based guideline for the assessment and management of polycystic ovary syndrome. Hum Reprod 2018;33:1602-18. [Crossref] [PubMed]
- Balen AH, Morley LC, Misso M, et al. The management of anovulatory infertility in women with polycystic ovary syndrome: an analysis of the evidence to support the development of global WHO guidance. Hum Reprod Update 2016;22:687-708. [Crossref] [PubMed]
- Pfieffer ML. Polycystic ovary syndrome: An update. Nursing 2019;49:34-40. [Crossref] [PubMed]
- Flippot R, Beinse G, Boilève A, et al. Long non-coding RNAs in genitourinary malignancies: a whole new world. Nat Rev Urol 2019;16:484-504. [Crossref] [PubMed]
- Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 2015;47:199-208. [Crossref] [PubMed]
- Carninci P, Kasukawa T, Katayama S, et al. The transcriptional landscape of the mammalian genome. Science 2005;309:1559-63. [Crossref] [PubMed]
- Katayama S, Tomaru Y, Kasukawa T, et al. Antisense Transcription in the Mammalian Transcriptome. Science 2005;309:1564-6. [Crossref] [PubMed]
- ENCODE Project Consortium, Birney E, Stamatoyannopoulos JA, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007;447:799-816. [Crossref] [PubMed]
- Schmitz SU, Grote P, Herrmann BG. Mechanisms of Long Noncoding RNA Function in Development and Disease. Cell Mol Life Sci 2016;73:2491-509. [Crossref] [PubMed]
- Kung JT, Colognori D, Lee JT. Long Noncoding RNAs: Past, Present, and Future. Genetics 2013;193:651-69. [Crossref] [PubMed]
- Wilusz JE, Freier SM, Spector DL. 3’ end processing of a long nuclear-retained non coding RNA yields at RNA-like cytoplasmic RNA. Cell 2008;135:919-32. [Crossref] [PubMed]
- Yin QF, Yang L, Zhang Y, et al. Long noncoding RNAs with snoRNA ends. Mol Cell 2012;48:219-30. [Crossref] [PubMed]
- Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007;316:1484-8. [Crossref] [PubMed]
- Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol 2013;20:300-7. [Crossref] [PubMed]
- Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 2016;17:47-62. [Crossref] [PubMed]
- Cruz JA, Westhof E. The dynamic landscapes of RNA architecture. Cell 2009;136:604-9. [Crossref] [PubMed]
- Tsai MC, Manor O, Wan Y, et al. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science 2010;329:689-93. [Crossref] [PubMed]
- Brannan CI, Dees EC, Ingram RS, et al. The product of the H19 gene may function as an RNA. Mol Cell Biol 1990;10:28-36. [Crossref] [PubMed]
- Hüttenhofer A, Schattner P, Polacek N. Non-coding RNAs: Hope or hype? Trends Genet 2005;21:289-97. [Crossref] [PubMed]
- Dinger ME, Amaral PP, Mercer TR, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res 2008;18:1433-45. [Crossref] [PubMed]
- Mercer TR, Dinger ME, Sunkin SM, et al. Specific expression of long noncoding RNAs in the adult mouse brain. Proc Natl Acad Sci USA 2008;105:716-21. [Crossref] [PubMed]
- Wang KC, Chang HY. Molecular Mechanisms of Long Noncoding RNAs. Mol Cell 2011;43:904-14. [Crossref] [PubMed]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet 2009;10:155-9. [Crossref] [PubMed]
- Mercer TR, Mattick JS. Structure and Function of Long Noncoding RNAs in Epigenetic Regulation. Nat Struct Mol Biol 2013;20:300-7. [Crossref] [PubMed]
- Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature 2012;482:339-46. [Crossref] [PubMed]
- Zhao J, Sun BK, Erwin JA, et al. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 2008;322:750-6. [Crossref] [PubMed]
- Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007;129:1311-23. [Crossref] [PubMed]
- Lanz RB, Razani B, Goldberg AD, et al. Distinct RNA motifs are important for coactivation of steroid hormone receptors by steroid receptor RNA activator (SRA). Proc Natl Acad Sci USA 2002;99:16081-6. [Crossref] [PubMed]
- Hanly DJ, Esteller M, Berdasco M. Interplay between long non-coding RNAs and epigenetic machinery:emerging targets in cancer? Philos Trans R Soc Lond B Biol Sci 2018;373:20170074. [Crossref] [PubMed]
- Martianov I, Ramadass A, Barros S, et al. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 2007;445:666-70. [Crossref] [PubMed]
- Bernard D, Prasanth KV, Tripathi V, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J 2010;29:3082-93. [Crossref] [PubMed]
- Tripathi V, Ellis JD, Shen Z, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 2010;39:925-38. [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- Velthut-Meikas A, Simm J, Tuuri T, et al. Research resource: small RNA-seq of human granulosa cells reveals miRNAs in FSHR and aromatase genes. Mol Endocrinol 2013;27:1128-41. [Crossref] [PubMed]
- Wilhelmm D, Bernard P. Non-coding RNAs and the Reproductive System. Adv Exp Med Biol 2016;886:V. -Vi. [Crossref] [PubMed]
- Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells:functional role of a nuclear noncoding RNA. Mol Cell 2009;35:467-78. [Crossref] [PubMed]
- Yamazaki T, Hirose T. The building process of the functional paraspeckle with long non-coding RNAs. Front Biosci (Elite Ed) 2015;7:1-41. [Crossref] [PubMed]
- Nakagawa S, Shimada M. The lncRNA Neat1 is required forcorpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development 2014;141:4618-27. [Crossref] [PubMed]
- Qin C, Xia X, Fan Y, et al. A novel, noncoding-RNA-mediated, post-transcriptional mechanism of anti-Mullerian hormone regulation by the H19/let-7 axis. Biol Reprod 2019;100:101-11. [Crossref] [PubMed]
- Reddy P, Zheng W, Liu K. Mechanisms maintaining the dormancy and survival of mammalian primordial follicles. Trends Endocrinol Metab 2010;21:96-103. [Crossref] [PubMed]
- Binelli M, Murphy BD. Coordinated regulation of follicle development by germ and somatic cells. Reprod Fertil Dev 2010;22:1-12. [Crossref] [PubMed]
- Bouckenheimer J, Fauque P, Lecellier CH, et al. Differential long non-coding RNA expression profiles in human oocytes and cumulus cells. Sci Rep 2018;8:2202. [Crossref] [PubMed]
- Jiao J, Shi B, Wang TR, et al. Characterization of long non-coding RNA and messenger RNA profiles in follicular fluid from mature and immature ovarian follicles of healthy women and women with polycystic ovary syndrome. Hum Reprod 2018;33:1735-48. [Crossref] [PubMed]
- Ernst EH, Nielsen J, Ipsen MB, et al. Transcriptome Analysis of Long Non-coding RNAs and Genes Encoding Paraspeckle Proteins During Human Ovarian Follicle Development. Front Cell Dev Biol 2018;6:78. [Crossref] [PubMed]
- Reig-Viader R, Vila-Cejudo M, Vitelli V, et al. Telomeric Repeat-Containing RNA (TERRA) and Telomerase Are Components of Telomeres During Mammalian Gametogenesis. Biol Reprod 2014;90:103. [PubMed]
- Reig-Viader R, Brieño-Enríquez MA, Khoriauli L, et al. Telomeric repeat-containing RNA and telomerase in human fetal oocytes. Hum Reprod 2013;28:414-22. [Crossref]
- Sang X, Zhang YZ. Long Non-coding RNA NEAT1 Drives the Development of Polycystic Ovary Syndrome via Sponging Multiple MicroRNA. Cell Biol Int 2020. Epub ahead of print. [Crossref] [PubMed]
- Zhu HL, Chen YQ, Zhang ZF. Downregulation of lncRNA ZFAS1 and upregulation of microRNA-129 repress endocrine disturbance, increase proliferation and inhibit apoptosis of ovarian granulosa cells in polycystic ovarian syndrome by downregulating HMGB1. Genomics 2020;112:3597-608. [Crossref] [PubMed]
- Zhang D, Yuan TH, Tan L, et al. MALAT1 is involved in the pathophysiological process of PCOS by modulating TGFβ signaling in granulosa cells. Mol Cell Endocrinol 2020;499:110589. [Crossref] [PubMed]
- Liu G, Liu S, Xing G, et al. LncRNA PVT1/MicroRNA-17-5p/PTEN Axis Regulates Secretion of E2 and P4, Proliferation, and Apoptosis of Ovarian Granulosa Cells in PCOS. Mol Ther Nucleic Acids 2020;20:205-16. [Crossref] [PubMed]
- Chen Y, Zhang X, An Y, et al. LncRNA HCP5 promotes cell proliferation and inhibits apoptosis via miR27a-3p/IGF-1 axis in human granulosa-like tumor cell line KGN. Mol Cell Endocrinol 2020;503:110697. [Crossref] [PubMed]
- Yang R, Chen J, Wang L, et al. LncRNA BANCR participates in polycystic ovary syndrome by promoting cell apoptosis. Mol Med Rep 2019;19:1581-6. [PubMed]
- Zhao J, Xu JY, Wang WS, et al. Long non-coding RNA LINC-01572:28 inhibits granulosa cell growth via a decrease in p27 (Kip1) degradation in patients with polycystic ovary syndrome. EBioMedicine 2018;36:526-38. [Crossref] [PubMed]
- Liu YD, Li Y, Feng SX, et al. Long Noncoding RNAs: Potential Regulators Involved in the Pathogenesis of Polycystic Ovary Syndrome. Endocrinology 2017;158:3890-9. [Crossref] [PubMed]
- Wu G, Yang Z, Chen Y, et al. Downregulation of Lnc-OC1 attenuates the pathogenesis of polycystic ovary syndrome. Mol Cell Endocrinol 2020;506:110760. [Crossref] [PubMed]
- Che Q, Liu M, Zhang DD, et al. Long Noncoding RNA HUPCOS Promotes Follicular Fluid Androgen Excess in PCOS Patients via Aromatase Inhibition. J Clin Endocrinol Metab 2020;105:1-12. [Crossref] [PubMed]
- Zhao J, Huang J, Geng X, et al. Polycystic Ovary Syndrome: Novel and Hub lncRNAs in the Insulin Resistance-associated lncRNA-mRNA Network. Front Genet 2019;10:772. [Crossref] [PubMed]
- Huang X, Hao C, Bao H, et al. Aberrant expression of long noncoding RNAs in cumulus cells isolated from PCOS patients. J Assist Reprod Genet 2016;33:111-21. [Crossref] [PubMed]
- Huang X, Pan JP, Wu B, et al. Construction and analysis of a lncRNA (PWRN2)-mediated ceRNA network reveal its potential roles in oocyte nuclear maturation of patients with PCOS. Reprod Biol Endocrinol 2018;16:73. [Crossref] [PubMed]
- Butler AE, Hayat S, Dargham SR, et al. Alterations in long noncoding RNAs in women with and without polycystic ovarian syndrome. Clin Endocrinol (Oxf) 2019;91:793-7. [Crossref] [PubMed]
- Liu Z, Hao C, Huang X, et al. Peripheral blood leukocyte expression level of lncRNA steroid receptor RNA activator (SRA) and its association with polycystic ovary syndrome: a case control study. Gynecol Endocrinol 2015;31:363-8. [Crossref] [PubMed]
- Liu Z, Hao C, Song D, et al. Androgen Receptor Coregulator CTBP1-AS Is Associated With Polycystic Ovary Syndrome in Chinese Women: A Preliminary Study. Reprod Sci 2015;22:829-37. [Crossref] [PubMed]
- Lin H, Xing W, Li Y, et al. Downregulation of serum long noncoding RNA GAS5 may contribute to insulin resistance in PCOS patients. Gynecol Endocrinol 2018;34:784-8. [Crossref] [PubMed]
- Qin L, Huang CC, Yan XM, et al. Long non-coding RNA H19 is associated with polycystic ovary syndrome in Chinese women: A preliminary study. Endocr J 2019;66:587-95. [Crossref] [PubMed]
- Liu M, Zhu HQ, Li Y, et al. Expression of serum lncRNA-Xist in patients with polycystic ovary syndrome and its relationship with pregnancy outcome. Taiwan J Obstet Gynecol 2020;59:372-6. [Crossref] [PubMed]
- Li L, Zhu J, Ye F, et al. Upregulation of the lncRNA SRLR in polycystic ovary syndrome regulates cell apoptosis and IL-6 expression. Cell Biochem Funct 2020;38:880-5. [Crossref] [PubMed]
- Chang HM, Qiao J, Leung PC. Oocyte–somatic cell interactions in the human ovary—novel role of bone morphogenetic proteins and growth differentiation factors. Hum Reprod Update 2016;23:1-18. [Crossref] [PubMed]
- Eißmann M, Gutschner T, Hämmerle M, et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol 2012;9:1076-87. [Crossref] [PubMed]
- Jin L, Yang Q, Zhou CL, et al. Profiles for long non-coding RNAs in ovarian granulosa cells from women with PCOS with or without hyperandrogenism. Reprod Biomed Online 2018;37:613-23. [Crossref] [PubMed]
- Sang Q, Yao Z, Wang H, et al. Identification of microRNAs in human follicular fluid: characterization of microRNAs that govern steroidogenesis in vitro and are associated with polycystic ovary syndrome in vivo. J Clin Endocrinol Metab 2013;98:3068-79. [Crossref] [PubMed]
- Machtinger R, Laurent LC, Baccarelli AA. Extracellular vesicles: roles in gamete maturation, fertilization and embryo implantation. Hum Reprod Update 2016;22:182-93. [PubMed]
- Walters KA, Bertoldo MJ, Handelsman DJ. Evidence from animal models on the pathogenesis of PCOS. Best Pract Res Clin Endocrinol Metab 2018;32:271-81. [Crossref] [PubMed]
- Jiang B, Xue M, Xu DB, et al. Down-regulated lncRNA HOTAIR alleviates polycystic ovaries syndrome in rats by reducing expression of insulin-like growth factor 1 via microRNA-130a. J Cell Mol Med 2020;24:451-64. [Crossref] [PubMed]
- Zhang X, Xu Y, Fu L, et al. Identification of mRNAs related to endometrium function regulated by lncRNA CD36-005 in rat endometrial stromal cells. Reprod Biol Endocrinol 2018;16:96. [Crossref] [PubMed]
- Fu LL, Xu Y, Li DD, et al. Expression profiles of mRNA and long noncoding RNA in the ovaries of letrozoleinduced polycystic ovary syndrome rat model through deep sequencing. Gene 2018;657:19-29. [Crossref] [PubMed]
- Li Y, Zhao W, Wang H, et al. Silencing of LncRNA Steroid Receptor RNA Activator attenuates polycystic ovary syndrome in Mice. Biochimie 2019;157:48-56. [Crossref] [PubMed]


