
Effect of activation of the Akt/mTOR signaling pathway by EEF1A2 on the biological behavior of osteosarcoma
Introduction
Osteosarcoma (OS) is the most common primary bone cancer in children and adolescents, and it causes a large number of cancer-related deaths worldwide every year (1,2). Although extensive tumor resection combined with adjuvant chemotherapy and radiotherapy has allowed significant progress in the treatment of OS, the clinical prognosis of OS patients is still poor, due to its characteristics of high metastatic potential and rapid progress (3,4). Therefore, there is an urgent need to study the molecular mechanism of OS occurrence and development to lay the foundation for developing new effective therapeutic targets.
Eukaryotic Translation Elongation Factor 1 Alpha 2 (EEF1A2) is selectively expressed by the heart, skeletal muscle, nervous system, and some special cells. It plays an important biological role by promoting the guanosine triphosphate (GTP)-dependent binding of aminoacyl tRNA to ribosomal A sites during protein biosynthesis (5,6). The abnormal expression of EEF1A2 is closely related to many diseases, such as Parkinson's disease and epilepsy (7,8). Increasingly, research has focused on the role of EEF1A2 in tumors, and it has been revealed as overexpressed in many human tumors, to have carcinogenic properties, and promote tumor progression. The role of its oncogene has been verified in most tumors, including colorectal cancer, leukemia, breast cancer, hepatocellular carcinoma, and gastric cancer, among others (9-13). However, there is no research on EEF1A2 in OS, and its expression and role in OS are still unknown.
Therefore, in this study, we aimed to explore the role of EEF1A2 in OS. First, through bioinformatics analysis, it was found that EEF1A2 was highly expressed in OS. Subsequently, OS tissues and cell lines were used to verify its expression and analyze its clinical significance. Furthermore, the effects of EEF1A2 on the proliferation, migration, and invasion of OS cell lines in vitro and the growth of OS tumors in vivo were investigated. Finally, the tumor-related signaling pathway regulated by EEF1A2 was demonstrated, and the mechanism of its biological role was explained. We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-7974).
Methods
Tissue sample collection
In this study, 38 OS specimens and matched normal tissue specimens that had been surgically removed in our hospital were collected. The clinical parameters of OS patients are shown in Table 1. All participants in this study provided written informed consent. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the ethics committee of our hospital and conducted in accordance with the guidelines provided by the ethics committee and the institutional review committee (No. 2018087).
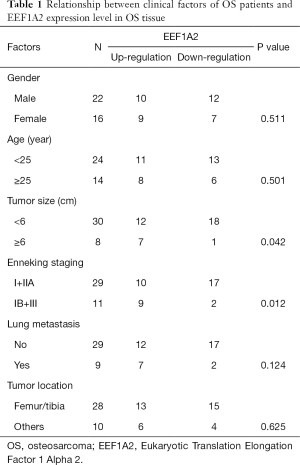
Full table
Cell culture and transfection
Human OS cell lines (MG63, HOS, U2OS, and Saos-2) and human osteoblast cell lines (hFOB1.19) were purchased from ATCC (American Type Culture Collection, Manassas, VA, USA). The cells were cultured in Dulbecco’s modified eagle medium (DMEM) (HyClone, Logan, UT, USA) containing 10% fetal bovine serum (FBS, Gibco, Gaithersburg, MD, USA), 100 U/mL penicillin, and 100 mg/mL streptomycin (Invitrogen, Waltham, MA, USA). All cell lines were incubated in an incubator containing 5% CO2 at 37 °C. Lipofectamine 2000 (Invitrogen) was used for cell transfection, and the transfection procedure was performed according to the manufacturer instructions.
RNA extraction and real-time fluorescence quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Total RNA was extracted with Trizol reagent (Invitrogen) from tissue or cell, and then DNA was removed with TurboDNase Kit (Ambion, Austin, TX, USA). The total RNA of each sample was quantified using an ND-1000 spectrophotometer (NanoDrop, Wilmington, DE, USA), and the purity of RNA was evaluated using an absorbance ratio (A260/A280) of 260:280 nm. Thereafter, the extracted total RNA was reverse transcribed into complementary (c)DNA using Primer Script RT Mix kit (Takara, Shiga, Japan), and PCR was performed on the ABI PRISM 7500 system (Applied Biosystems, Foster City, CA, USA) using SYBR Green PCR kit (Takara). The glyceraldehyde 3-phosphate dehydrogenase (GAPDH) level was used as an internal reference, and the relative expression of EEF1A2 was calculated according to the 2−△△Ct method (13).
Cell proliferation experiment
A cell counting kit-8 (CCK-8) experiment and 5-ethynyl-2’-deoxyuridine (EdU) experiment were used to study cell proliferation. In the CCK-8 experiment, cells were seeded in 96-well plates (2,000 cells/well) and transfected. The CCK-8 reagent (Dojindo, Kumamoto, Japan) was added at 24, 48, and 72 hours after transfection, and the absorbance was measured at 450 nm with a spectrophotometer. In the EdU experiment, EdU cell proliferation assay (Sigma-Aldrich, St. Louis, MO, USA) was used to stain the cells with EdU and photographed with a fluorescence microscope. The EdU and 5-TAMRA-PEG3-Azide positive rate represented the relative proliferative capacity of the cells.
Cell migration and invasion experiments
Scratch tests were used to study the invasive ability of cells. After transfection, the cells were seeded in a 6-well plate at 100% density, and a scratch was made with the tip of a 10 µL pipette. Then, the scratch line was photographed at 0 and 24 hours. The healing ability of the scratch was measured by calculating the distance between the 2 sides of the scratch. The scratch width at 0 hours was used as a control to calculate the relative cell mobility.
Transwell cells (Corning Incorporated, Corning, NY, USA) with an aperture of 8 mm were used to study cell invasion.
First, 1×105 cells were inoculated with serum-free DMEM in the upper chamber of Transwell coated with matrix gel, and the lower chamber was added with complete medium containing 10% FBS. After 24 hours of cell culture, the upper chamber cells were removed with a cotton swab, while other cells were fixed with 4% paraformaldehyde, and stained with 0.5% crystal violet solution. Finally, the number of cells penetrating the membrane was counted under the microscope.
Nude mouse xenograft tumor model
Male Bagg albino (BALB)/c nude mice aged 4–6 weeks were used for in vivo xenograft studies. All animal experiments were approved by the Animal Ethics Committee of this hospital and in compliance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The nude mice were injected subcutaneously with MG63 cells with silenced EEF1A2 in the exponential growth phase. After that, the tumor size was measured with a caliper to calculate the tumor volume every 5 days. The mice were sacrificed 20 days after the tumor cells were inoculated, and the tumor weight was measured.
Western blot experiment
Total protein was extracted from cells with radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime, Shanghai, China), and protein concentration was determined with the bicinchoninic acid (BCA) protein quantification kit (BioRad, Hercules, CA, USA). First, an equal amount of protein (20 mg) was subjected to sodium dodecyl sulfate polyacrylamide gel (SDS-PAGE) electrophoresis and then transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, Burlington, MA, USA). After blocking with 5% skimmed milk powder for 1 hour at room temperature, it was incubated with the primary antibody (Abcam, Cambridge, MA, USA) at 4 °C overnight. The membrane was then washed with tris buffered saline with Tween 20 (TBST) and incubated with horseradish peroxidase (HRP)-conjugated secondary antibody at room temperature for 1 hour. Finally, the signals were detected using the enhanced chemiluminescence (ECL) solution (ABP Biosciences, Rockville, MD, USA) and a chemiluminescence imaging system (BioRad).
Statistical analysis
The data were expressed in the form of mean ± standard deviation. Each experiment was repeated at least 3 times and analyzed using the software SPSS version 19.0 (IBM, Armonk, NY, USA). The Student’s t-test and one-way analysis of variance (ANOVA) were used to analyze the differences between groups. Statistical significance was considered at P<0.05.
Results
Analysis of EEF1A2 expression in OS using GEO database
First, the gene expression of 5 normal vs. 6 tumors in the Gene Expression Omnibus (GEO) database (GSE126209_Osteosarcoma) were analyzed and we found that EEF1A2 had the most significant change among all genes. The threshold of differentially expressed genes was P<0.05, and the absolute value of log2-fold change (FC) was greater than 2; the gene labeling threshold was P<0.01, and log2-FC was greater than 5.5 (Figure 1A). Analysis of the expression of EEF1A2 revealed that EEF1A2 was significantly overexpressed in OS (Figure 1B).
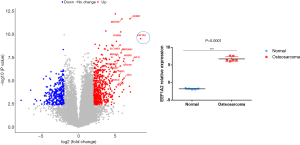
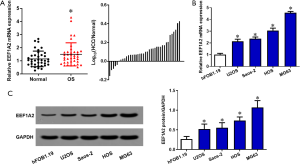
There is high expression of EEF1A2 in OS tissues and cell lines
To verify the expression of EEF1A2 in OS, qRT-PCR was used to detect the expression of EEF1A2 in OS tissues and cell lines. The results showed that in 38 OS tissues and paired normal tissues, the expression of EEF1A2 in OS tissue was significantly higher than that of normal bone tissue (Figure 2A). The expression of EEF1A2 in all OS cell lines (MG63, HOS, U2OS, and Saos-2) was higher than that of human osteoblast cell lines (hFOB1.19) (Figure 2B). After comparing with the clinical data of the patients, it was found that the expression of EEF1A2 was closely related to the Enneking stage of OS and tumor size (Table 1).
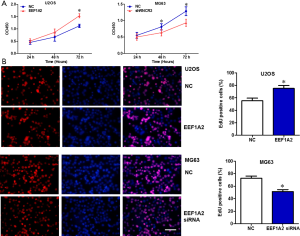
Cell proliferation of OS is promoted by EEF1A2 in vitro
After the transfection of EEF1A2 overexpression plasmid in U2OS cells, the results of the CCK8 experiment and clone formation experiment showed that the proliferation ability of the cells was significantly improved (Figure 3). However, after silencing EEF1A2 by the transfection of EEF1A2 small interfering (si)RNA in MG63 cells, the results of the CCK8 experiment and clone formation experiment demonstrated that the proliferation ability of the cells was significantly reduced (Figure 3). These results indicated that EEF1A2 promoted OS cell proliferation in vitro.
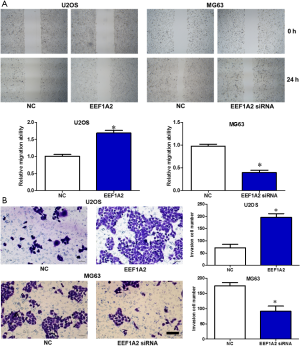
EEF1A2 promotes OS cell migration and invasion in vitro
After the transfection of EEF1A2 expression plasmid in U2OS cells, Transwell experiment results showed that the migration and invasion ability of cells was significantly improved (Figure 4). However, after silencing EEF1A2 by the transfection of EEF1A2 siRNA in MG63 cells, Transwell experiments showed that the migration and invasion ability of cells was significantly reduced (Figure 4). These results indicated that EEF1A2 can promote OS cell migration and invasion in vitro.
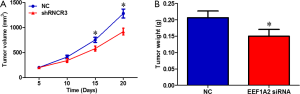
EEF1A2 promotes OS tumor growth in vivo
Compared with the control group, the growth rate of xenografts in nude mice constructed with MG63 cells that stably silence EEF1A2 was significantly slower (Figure 5A). The weight of the transplanted tumor in the EEF1A2 silent group was significantly lower than that in the control group (Figure 5B). These results indicated that EEF1A2 can promote the growth of OS tumors in vivo.
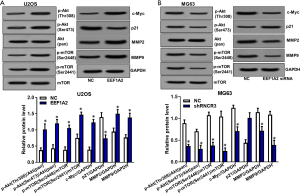
EEF1A2 activates Akt/mTOR signaling pathway in OS
After the transfection of EEF1A2 overexpression plasmid in U2OS cells, western blot experiments showed that the phosphorylation levels of protein kinase B (Akt) and mammalian target of rapamycin (mTOR) were significantly increased (Figure 6). However, after silencing EEF1A2 by the transfection of EEF1A2 siRNA in MG63 cells, western blot experiments showed that the phosphorylation levels of Akt and mTOR were significantly reduced (Figure 6). These results indicated that EEF1A2 can activate the Akt/mTOR signaling pathway in OS. We also showed that the proliferation, invasion and migration related proteins downstream Akt/mTOR signaling pathway including c-Myc, p21, MMP2 and MMP9 we regulated by EEF1A2 (Figure 6).
Discussion
Anand et al. (14) first reported the role of EEF1A2 in tumors and found that EEF1A2 was amplified in 25% of primary ovarian tumors and highly expressed in approximately 30% of ovarian tumors and established cell lines, showing that EEF1A2 has carcinogenic properties in vivo and in vivo and is a possible oncogene for ovarian cancer. Subsequently, increasing numbers of studies have shown that EEF1A2 is significantly overexpressed in various cancers (9-13). It is reported that EEF1A2 is closely related to the occurrence and development of tumors and poor prognosis. For example, in pancreatic cancer, high expression of EEF1A2 is associated with lymph node metastasis, peripheral nerve invasion, and poor prognosis (15); EEF1A2 can also promote the expression and activity of matrix metallopeptidase 9 (MMP-9), thereby promoting the migration and invasion of pancreatic cancer cells (16). In hepatocellular carcinoma, EEF1A2 mRNA and protein levels are significantly higher than those in matched adjacent tissues and normal tissues, while silencing EEF1A2 can lead to reduced cell proliferation, migration, and invasion, increased apoptosis, and induced cell cycle arrest (12). Moreover, many scholars believe that EEF1A2 is an important target for cancer treatment and expect it to be used clinically (6,17).
Because EEF1A2, as an oncogene, affects the occurrence and development of tumors, and its role in OS is unknown, in this study we explored whether EEF1A2 also plays the same role in OS. Through bioinformatics analysis, we found that EEF1A2 was significantly up-regulated in OS. Next, the expression of EEF1A2 in OS tissues and cell lines were examined, and we discovered that EEF1A2 expression was significantly increased in OS tissues and cell lines, and its high expression was closely related to OS Enneking stage and tumor size. Through subsequent functional studies, it was found that EEF1A2 can significantly promote the proliferation, migration, and invasion of OS cells in vitro, and the growth of OS tumors in vivo. Therefore, we confirmed that EEF1A2 is also highly expressed in OS, as an oncogene to promote tumorigenesis and development. Since there has not been any research on EEF1A2 in OS in the past, this study supplemented knowledge of the biological function of EEF1A2 and was very innovative.
At the same time, we also found that EEF1A2 can activate the Akt/mTOR signaling pathway in OS, explaining the mechanism of EEF1A2 exerting its biological function. It has been reported that EEF1A2 induces the activation of the Akt proto-oncogene in liver cancer, pancreatic cancer, and rat plasmacytoma (16,18,19). Here, we demonstrated that EEF1A2 can promote the phosphorylation of Akt and mTOR in OS and activate the Akt/mTOR signaling pathway. The Akt/mTOR signaling pathway is commonly active where tumors are present, and is involved in tumor cell metabolism, autophagy, proliferation, survival, invasion, and migration (20,21). We also showed that the proliferation, invasion and migration related proteins downstream Akt/mTOR signaling pathway including c-Myc, p21, MMP2 and MMP9 we regulated by EEF1A2. Therefore, we believe that EEF1A2 promotes the proliferation, migration, and invasion of OS cells in vitro and the growth of OS tumors in vivo by activating the Akt/mTOR signaling pathway. Beyond that, in other cancers, eEF1A2 has been reported to activate other cancer related signaling pathways. For instance, eEF1A2 is highly expressed in HCC and likely promote HCC tumorigenesis by activating PI3K/Akt/NF-κB signaling (12). Eef1a2 also promotes cell growth, inhibits apoptosis and activates JAK/STAT in mouse plasmacytomas (19). We suspect that eEF1A2 may also facilitate OS tumorigenesis by activating these signaling pathways.
Acknowledgments
Funding: This research was funded by the Sichuan Provincial Key R&D projects (20ZDYF1537).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-7974
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-7974
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-7974). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the ethics committee of our hospital and conducted in accordance with the guidelines provided by the ethics committee and the institutional review committee (No. 2018087). All animal experiments were approved by the Animal Ethics Committee of this hospital and in compliance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Scotlandi K, Hattinger CM, Pellegrini E, et al. Genomics and Therapeutic Vulnerabilities of Primary Bone Tumors. Cells 2020;9:968. [Crossref] [PubMed]
- Whelan JS, Davis LE. Osteosarcoma, Chondrosarcoma, and Chordoma. J Clin Oncol 2018;36:188-93. [Crossref] [PubMed]
- Carina V, Costa V, Sartori M, et al. Adjuvant Biophysical Therapies in Osteosarcoma. Cancers (Basel) 2019;11:348. [Crossref] [PubMed]
- Takeuchi A, Yamamoto N, Hayashi K, et al. Joint-preservation surgery for pediatric osteosarcoma of the knee joint. Cancer Metastasis Rev 2019;38:709-22. [Crossref] [PubMed]
- Long K, Wang H, Song Z, et al. EEF1A2 mutations in epileptic encephalopathy/intellectual disability: Understanding the potential mechanism of phenotypic variation. Epilepsy Behav 2020;105:106955. [Crossref] [PubMed]
- Losada A, Muñoz-Alonso MJ, García C, et al. Translation Elongation Factor eEF1A2 is a Novel Anticancer Target for the Marine Natural Product Plitidepsin. Sci Rep 2016;6:35100. [Crossref] [PubMed]
- Prommahom A, Dharmasaroja P. Effects of eEF1A2 knockdown on autophagy in an MPP+-induced cellular model of Parkinson's disease. Neurosci Res 2020. Epub ahead of print. [Crossref] [PubMed]
- Carvill GL, Helbig KL, Myers CT, et al. Damaging de novo missense variants in EEF1A2 lead to a developmental and degenerative epileptic-dyskinetic encephalopathy. Hum Mutat 2020;41:1263-79. [Crossref] [PubMed]
- Shen Z, Li Y, Fang Y, et al. SNX16 activates c-Myc signaling by inhibiting ubiquitin-mediated proteasomal degradation of eEF1A2 in colorectal cancer development. Mol Oncol 2020;14:387-406. [Crossref] [PubMed]
- Dapas B, Pozzato G, Zorzet S, et al. Effects of eEF1A1 targeting by aptamer/siRNA in chronic lymphocytic leukaemia cells. Int J Pharm 2020;574:118895. [Crossref] [PubMed]
- Giudici F, Petracci E, Nanni O, et al. Elevated levels of eEF1A2 protein expression in triple negative breast cancer relate with poor prognosis. PLoS One 2019;14:e0218030. [Crossref] [PubMed]
- Qiu FN, Huang Y, Chen DY, et al. Eukaryotic elongation factor-1α 2 knockdown inhibits hepatocarcinogenesis by suppressing PI3K/Akt/NF-κB signaling. World J Gastroenterol 2016;22:4226-37. [Crossref] [PubMed]
- Yang S, Lu M, Chen Y, et al. Overexpression of eukaryotic elongation factor 1 alpha-2 is associated with poorer prognosis in patients with gastric cancer. J Cancer Res Clin Oncol 2015;141:1265-75. [Crossref] [PubMed]
- Anand N, Murthy S, Amann G, et al. Protein elongation factor EEF1A2 is a putative oncogene in ovarian cancer. Nat Genet 2002;31:301-5. [Crossref] [PubMed]
- Duanmin H, Chao X, Qi Z. eEF1A2 protein expression correlates with lymph node metastasis and decreased survival in pancreatic ductal adenocarcinoma. Hepatogastroenterology 2013;60:870-5. [PubMed]
- Xu C, Hu DM, Zhu Q. eEF1A2 promotes cell migration, invasion and metastasis in pancreatic cancer by upregulating MMP-9 expression through Akt activation. Clin Exp Metastasis 2013;30:933-44. [Crossref] [PubMed]
- Losada A, Muñoz-Alonso MJ, Martínez-Díez M, et al. Binding of eEF1A2 to the RNA-dependent protein kinase PKR modulates its activity and promotes tumour cell survival. Br J Cancer 2018;119:1410-20. [Crossref] [PubMed]
- Pellegrino R, Calvisi DF, Neumann O, et al. EEF1A2 inactivates p53 by way of PI3K/AKT/mTOR-dependent stabilization of MDM4 in hepatocellular carcinoma. Hepatology 2014;59:1886-99. [Crossref] [PubMed]
- Li Z, Qi CF, Shin DM, et al. Eef1a2 promotes cell growth, inhibits apoptosis and activates JAK/STAT and AKT signaling in mouse plasmacytomas. PLoS One 2010;5:e10755. [Crossref] [PubMed]
- Xu F, Na L, Li Y, et al. Roles of the PI3K/AKT/mTOR signalling pathways in neurodegenerative diseases and tumours. Cell Biosci 2020;10:54. [Crossref] [PubMed]
- Ortega MA, Fraile-Martínez O, Asúnsolo Á, et al. Signal Transduction Pathways in Breast Cancer: The Important Role of PI3K/Akt/mTOR. J Oncol 2020;2020:9258396. [Crossref] [PubMed]
(English Language Editor: J. Jones)


