
The roles and mechanisms of the circular RNA circ_104640 in early-stage lung adenocarcinoma: a potential diagnostic and therapeutic target
Introduction
Globally, lung cancer (LC) remains the major cause of cancer deaths, accounting for approximately 13.2% of new cancer cases and 25.9% of cancer-related deaths (1). Non-small-cell lung cancer (NSCLC) is the main type of LC (85%), and can be subdivided into adenocarcinoma, squamous cell carcinoma, and large cell carcinoma (2). In China, the incidence of lung adenocarcinoma (LUAC) has surpassed that of lung squamous cell carcinoma (LSCC) to become the main type of NSCLC (3). Around 16% of LUAC cases among non-smokers in China are potentially attributable to passive smoking, particularly in women (about 18%). Most cases are the result of household exposure (4). Although novel treatments have significantly improved outcomes, the 5-year survival rate of LUAC patients is about 71–83%. For early-stage LUAC patients, surgery is the first choice for treatment (5). However, the incidence of recurrence is 21.7% (6). To facilitate early diagnosis and treatment, it is crucial to the understand the pathogenesis and identify the potential diagnostic markers of early-stage LUAC.
The early diagnosis of LUAC is largely dependent on imaging and laboratory tests. With the increasing use of low-dose computed tomography (LDCT), the LC mortality rate in men has decreased by 26%, and the mortality rate in the smaller female population has also decreased remarkably. It is worth noting that about 50% of diagnosed cancers are in the early stages (7). In recent years, the use of LDCT to detect early-stage LC has increased by 74.1% (8). Additionally, circulating tumor cells (CTCs), circulating tumor DNAs (ctDNAs), and exosomes have gradually become more vital in the early diagnosis of LUAC (9-11). Continuous progress is being made to develop novel early-stage LUAC diagnostic methods. The increased understanding of circular RNAs (circRNAs) in recent years has prompted researchers to explore its potential diagnostic, prognostic, predictive, and therapeutic value.
miRNAs (MicroRNAs) are small, non-coding RNA, typically 18–25 bases in length (12). miRNAs are involved in cancer cell proliferation, invasion and metastasis in LUAC (13,14). miRNAs usually bind to the 3'-untranslated region (UTR) of mRNA to silence its expression at the post-transcriptional level (12). The ligand-receptor pair CCL20 (C-C motif chemokine ligand 20) / CCR6 (C-C motif chemokine receptor 6) are involved in the metastasis of a variety of tumors, including LUAC and CRC (colorectal carcinomas) (15,16).
This investigation analyzed data from the Gene Expression Omnibus (GEO) database to identify the differentially expressed circRNAs, miRNAs, and messenger RNAs (mRNAs) in early-stage LUAC patients. The abnormal overexpression of these RNAs were verified in the tumor tissues of 35 early-stage LUAC patients. The circ_104640 was stably expressed and mainly enriched in the cytoplasm, and it could sponge miRNAs to promote the expression of CCL20. This report described a novel circ_104640 regulatory mechanism and may provide a basis for the diagnosis and treatment of early-stage LUAC.
We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/atm-20-8019).
Methods
Identification of the differentially expressed circRNAs, miRNAs, and mRNAs
The microarray data were download from the GEO database (http://www.ncbi.nlm.nih.gov/geo/) (17). The circRNA, miRNA, and mRNA expression profiles were obtained from the Genomic Spatial Event (GSE)101684, GSE135918, and GSE136043, respectively (18,19). The differentially expressed circRNAs, miRNAs, and mRNAs were screened using the GEO2R tool, with thresholds of |log2(FC)| >1.0 and adj. P<0.05. The data download from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/) and the Genotype-Tissue Expression (GTEx) database (https://www.gtexportal.org/) were analyzed using the above methods (20,21).
Clinical samples
This study included 35 patients with early-stage LUAC (stage I–II) who underwent surgical treatment at the Thoracic Surgery Department of the First Affiliated Hospital of Soochow University between September 2019 and May 2020. None of the patients suffered from other primary tumors or recurrent tumors and none had undergone radiotherapy or chemotherapy prior to surgery. Ethics approval was obtained (No.: 2018011) from the Ethics Committee of the First Affiliated Hospital of Soochow University and all patients provided signed informed consents. Tissue specimens were collected from the patients and stored in liquid nitrogen immediately after surgical resection. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Cell culture
The human LUAC cell lines, including A549, H1299, SPCA1, and PC9, were purchased from the Chinese Academy of Sciences (Shanghai, China) and cultivated in Dulbecco's Modified Eagle Medium (DMEM; Gibco, NY, USA) supplemented with 10% fetal bovine serum (FBS; Gibco) at 37 °C with 5% CO2.
Statistical analysis
All experiments were repeated in three times and data are expressed as the mean ± SD and analyzed using Student’s T-test or one-way ANOVA. The analysis was performed using GraphPad Prism 7.0 (Graphpad Software, CA, USA). Differences were considered significant if *P<0.05, **P<0.01, ***P<0.001, respectively.
More details can be found in the Supplementary file (Appendix 1).
Results
Circ_104640 is overexpressed in early-stage LUAC tissues
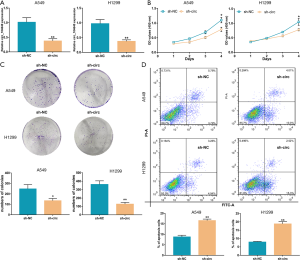
To clarify the role of circRNAs in early-stage LUAC, the microarray data set GSE101684, was analyzed. This microarray data contained the cancerous and adjacent normal lung samples of 4 patients with early-stage LUAC (3 stage IA patients and 1 stage IIA patient). Using the GEO2R online tool the top ten up-regulated and down-regulated circRNAs were identified (Figure 1A). Based on limitations of the length of circRNAs (300–1,000 bp), circ_101640 was selected for further research. As shown in Figure 1B, circ_104640, also known as circ_0001806, is derived from the centrosome and spindle pole associated protein 1 (CSPP1) gene, and is about 432 bp in length. Patients with tumor, node, metastasis (TNM) stage IA and IIA LUAC showed higher circ_104640 expression in their tumor tissues compared to the adjacent lung tissues (Figure 1C). Among the 35 patients with early-stage LUAC in our institution, this phenomenon was confirmed again, that is, the expression of circ_104640 in tumor tissues was significantly higher than that in the adjacent healthy tissues (Figure 1D). The levels of circ_104640 expression in LUAC cell lines were detected by qRT-PCR. The results demonstrated that circ_104640 was up-regulated most in the A549 and H1299 cells, compared with the SPCA1 and PC9 cells (Figure 1E). Further qRT-PCR analyses demonstrated that circ_104640 was resistant to RNase-R digestion. Addition of RNase-R reduced linear CSPP1 mRNA expression, but not circ_104640 expression (Figure 1F). The isolated RNA analysis of cytoplasmic and nuclear components showed that circ_104640 was mainly located in the cytoplasm of A549 and H1299 cells (Figure 1G). The above observations showed that circ_104640 mainly existed in the cytoplasm of LUAC cells and may serve as a promoter in the occurrence and progression of early-stage LUAC.

Circ_104640 accelerates proliferation and inhibits apoptosis in LUAC cells
To examine the biological role of circ_104640 in vitro, a short hairpin RNA (shRNA) and a negative control (NC) for circ_104640 were designed, namely sh-circ and sh-NC, respectively. Their efficacy was assessed in A549 and H1299 cells (Figure 2A). Cell counting kit 8 (CCK-8) assay revealed that silencing of circ_104640 by sh-circ resulted in a significant inhibition of cell viability and growth (Figure 2B). The colony formation assay showed that inhibition of circ_104640 impeded the ability of individual cells to form colonies (Figure 2C). In addition, as shown in Figure 2D, the proportion of apoptotic cells in the sh-circ group was significantly increased, demonstrating the ability of sh-circ to promote programmed cell death. Taken together, these data suggested that circ_104640 could accelerate cell proliferation and inhibit apoptosis in LUAC cells in vitro, thereby participating in the occurrence and progression of early-stage LUAC.
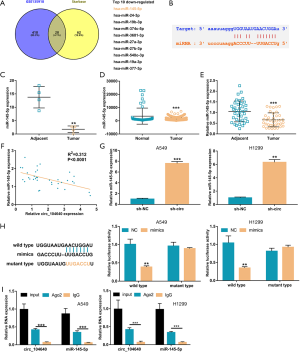
Circ_104640 serves as a sponge for miR-145-5p
Considering that circ_104640 was mainly detected in the cytoplasm of LUAC cells, and circRNAs contain binding sites that are complementary to miRNAs, they may act as miRNA “sponges” (22). To investigate this, the microarray data of GSE135918 were obtained from the GEO database and analyzed. The microarray data included miRNA expression in cancer tissues and adjacent non-tumor tissues of 4 early-stage LUAC patients (1 stage IA patient, 1 stage IB patient and 2 stage IIB patients). Based on the binding sites of circRNA-miRNA on the Starbase website, the Venn diagram analysis revealed the top ten down-regulated miRNAs, and miR-145-5p was selected for further investigation (Figure 3A,B). Previous studies have found that miR-145-5p expression in NSCLC tissues is markedly lower than that detected in healthy tissues (23). In agreement with this, the expression of miR-145-5p in the tumor tissues of the 4 early-stage LUAC patients was significantly reduced compared to the adjacent tissues (Figure 3C). In the TCGA LUAC database, a similar phenomenon was observed in tumor tissues compared to adjacent healthy tissues (Figure 3D). Similarly, in the 35 early-stage LUAC patients, miR-145-5p expression in the tumor tissues was decreased significantly compared to the healthy adjacent tissue (Figure 3E). In fact, in this study cohort, a negative correlation was found between circ_104640 and miR-145-5p expression (Figure 3F). Furthermore, the transfection of sh-circ reduced the expression of miR-145-5p in LUAC cells (Figure 3G). Subsequently, a dual luciferase reporter assay was conducted to identify the regulatory relationship between miR-145-5p and circ_104640. The miR-145-5p mimics reduced the luciferase activities of the wild-type circ_104640 vector, but had little effect on the mutant-type circ_104640 vector, indicating that miR-145-5p may be the target of circ_104640 (Figure 3H). The results of the argonaute 2 (Ago2) RNA immunoprecipitation (RIP) assay showed that RNA retrieval with the Ago2 antibody resulted in significantly increased levels of circ_104640 and miR-145-5p compared with the control (Figure 3I). These results indicated that miR-145-5p may function as a biomarker in early-stage LUAC and may be regulated by circ_104640.
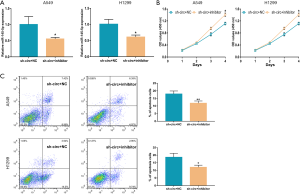
Circ_104640 may sponge miR-145-5p to regulate proliferation and apoptosis in vitro
The mechanisms by which circ_104640 and miR-145-5p exert their biological functions in LUAC cells were investigated. LUAC cell lines were transfected with the sh-circ, miR-145-5p inhibitor, or a NC, and the expression of miR-145-5p was assessed by qRT-PCR assay (Figure 4A). The CCK-8 assay demonstrated that the miR-145-5p inhibitor dampened the inhibitory effect of sh-circ on cell proliferation (Figure 4B). Flow cytometry studies revealed that the miR-145-5p inhibitor diminished the effects of sh-circ in LUAC cells and reduced the rate of apoptosis (Figure 4C). Therefore, our results showed that circ_104640 competitively combined with miR-145-5p in vitro to promote the proliferation and inhibit apoptosis of LUAC cells.
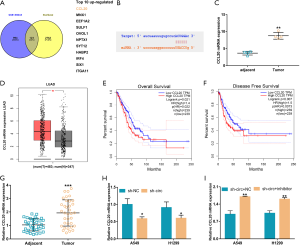
CCL20 may be the downstream target of miR-145-5p
To further explore the downstream regulatory mechanisms of miR-145-5p, the Starbase website and GSE136043 were analyzed. GSE136043 is the same batch of samples as GSE135918, which also includes 1 stage IA patient, 1 stage IB patient and 2 stage IIB patients. From the top ten up-regulated genes, CCL20 identified for further study (Figure 5A,B). In the tissue samples from the 4 patients with early-stage LUAC, the levels of CCL20 expression in the tumor tissues were markedly higher than that in non-tumor tissues (Figure 5C). In the TCGA LUAC database, by using the normal tissue calibration of the GTEx database, we found that the expression of CCL20 in tumor tissues was also significantly higher than that in normal tissues (Figure 5D). CCL20 expression was also closely related to the overall survival (OS) and disease-free survival (DFS) of LUAC patients, with statistical significance (Figure 5E,F). Again, in the tumor tissue samples of the 35 early-stage LUAC patients, the levels of CCL20 expression were remarkably increased in the tumor tissue compared to the adjacent healthy tissue (Figure 5G). In LUAC cells, the presence of sh-circ reduced the mRNA levels of CCL20, and this was partially restored in the presence of the inhibitor (Figure 5H,I). These results suggested that CCL20 is a potential target of miR-144-5p and can be influenced by circ_104640.
Discussion
Despite the wide applications of surgical techniques and targeted therapies, the 5-year survival rate of LUAC patients has not increased satisfactorily in recent years. Therefore, based on the principles of early diagnosis and early treatment, there is an urgency to better understand the biomolecular mechanisms facilitating the occurrence and development of early-stage LUAC.
CircRNAs are a type of non-coding RNA derived from back-splicing of the precursor mRNA. They have a covalently bonded, closed circular structure and a higher tolerance to exonuclease digestion (24). Due to the unique circular closed structure, high stability, and special expression patterns, it is believed that circRNAs can become potential biomarkers (25). In fact, circRNAs are dysregulated in various cancers, including LUACs (26-28). Several studies have suggested that dysregulated circRNAs in tissues or plasma could be used as novel biomarkers for cancer (29,30). Indeed, circ_0007385 has been shown to be upregulated in LUAC tissues and its expression was correlated with lymph node metastasis and TNM staging (31). Higher plasma circ_0000190 levels have been associated with larger tumors, worse histological types of adenocarcinoma, later stages of cancer, more distant metastatic organs, extra-thoracic metastasis, and poor survival and prognosis (32).
Up to now, only a few circRNAs have been reported in LUACs, especially in early-stage LUACs. Marked overexpression of circ_0001492, circ_0001346, circ_0000690 and circ_0001439 have been detected in exosomes in early-stage LUACs (33). Studies in the Chinese population have shown that the expression levels of circ_0047921, circ_0056285, and circ_0007761 could differentiate early-stage NSCLC cases from healthy controls, chronic obstructive pulmonary diseases, and tuberculosis (34). However, the roles of dysregulated circRNAs in early-stage LUAC is yet to be fully explored. Our studies revealed that circ_104640 is up-regulated in early-stage LUAC and may regulate cell proliferation and apoptosis.
In this report, GEO2R was used to select candidate circRNAs based on the research of other institutions. The expression of circ_104640 was abnormally high in early-stage LUAC tissues and it could exist stably in the cytoplasm. In vitro, interfering with the expression of circ_104640 significantly reduced the proliferative ability of LUAC cells, and increased cell apoptosis. These results suggested that circ_104640 may play a tumor-promoting role in the occurrence and progression of early-stage LUAC, and may represent a potential novel biomarker and therapeutic target in LUAC.
circRNAs have been identified as competing endogenous RNAs (ceRNAs) that reduce the expression of miRNAs that target mRNAs in tumors (22). To further study the mechanisms of circ_104640 in early-stage LUACs, the early-stage LUAC samples from another institution were analyzed to detect the expression of miRNAs. The expression of miR-145-5p was found to be down-regulated among the 4 stage I-II LUAC patients. This phenomenon was also observed in the 35 early-stage patients in our institution, and the expression level of miR-145-5p was negatively correlated with circ_104640. The potential interaction of circ_104640 and miR-145-5p was verified using the dual luciferase reporter assay and the Ago2 RIP assay. Moreover, we found that miR-145-5p inhibitors could reverse the biological functions of sh-circ in LUAC cells.
miRNAs also plays a key role in controlling cellular functions by repressing target genes (35). Chemokines are a superfamily of small chemotactic cytokines that regulate multiple biological functions by activating 7-transmembrane-domain G protein-coupled receptors (36). The C-C motif CCL20 is a member of the chemokines and can interact with the C-C motif chemokine receptor 6 (CCR6) in various diseases to facilitate tumor growth, angiogenesis, and escape antitumor immune surveillance through autocrine or paracrine pathways (37,38). Previous studies found that CCL20/CCR6 was closely related to the progression of LC. Both CCR6 and CCL20 were overexpressed in patients with recurrent LC, and CCL20 enhanced cell growth by activating extracellular signal-regulated kinase (ERK) signaling in A549 cells (39,40). The TCGA database showed that the expression of CCL20 was also markedly higher in tumor tissues compared to normal tissues and it was closely related to the OS and DFS of LUAC patients. Similarly, in this study, CCL20 was shown to be overexpressed in the early-stage LUAC tissues and was regulated by circ_104640 and miR-145-5p.
There were a number of limitations in this current study. The cohort size of the early-stage LUAC patients was small, and plasma samples were not collected. Therefore, we could not fully investigate the potential role of circ_104640 as a biomarker. Further sample collection will be required to demonstrate their potential as noninvasive biomarkers. In addition, the biological functions of other abnormally expressed circRNAs remain to be discovered. It is important to note that miR-145-5p is not the only target of circ_104640. Indeed, circ_104640 may regulate the progression of LUAC through sponging other miRNAs, and stabilizing or degrading other proteins, and this merits further investigation. Furthermore, the cell lines used in this study were obtained from advanced-stage LUAC patients, and hence could not provide a precise interpret of the mechanisms of early-stage LUAC occurrence and development.
In summary, this report provided new insights into the mechanisms of early-stage LUAC and suggested a novel target for the diagnosis and treatment of the disease.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (Grant no. 81672281 and 81800279), the Jiangsu Provincial Research Foundation for Basic Research (Grant no. BK20191174), and the Natural Science Foundation of Jiangsu Province (Grant no. BK20180197).
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/atm-20-8019
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-8019
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-8019). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Ethics approval was obtained (No.: 2018011) from the Ethics Committee of the First Affiliated Hospital of Soochow University and all patients provided signed informed consents. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Cronin KA, Ries LA, Edwards BK. The Surveillance, Epidemiology, and End Results (SEER) Program of the National Cancer Institute. Cancer 2014;120 Suppl 23:3755-7. [Crossref] [PubMed]
- Oser MG, Niederst MJ, Sequist LV, et al. Transformation from non-small-cell lung cancer to small-cell lung cancer: molecular drivers and cells of origin. Lancet Oncol 2015;16:e165-72. [Crossref] [PubMed]
- Yang D, Liu Y, Bai C, et al. Epidemiology of lung cancer and lung cancer screening programs in China and the United States. Cancer Lett 2020;468:82-7. [Crossref] [PubMed]
- Du Y, Cui X, Sidorenkov G, et al. Lung cancer occurrence attributable to passive smoking among never smokers in China: a systematic review and meta-analysis. Transl Lung Cancer Res 2020;9:204-17. [Crossref] [PubMed]
- Goldstraw P, Chansky K, Crowley J, et al. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J Thorac Oncol 2016;11:39-51. [Crossref] [PubMed]
- Wang C, Wu Y, Shao J, et al. Clinicopathological variables influencing overall survival, recurrence and post-recurrence survival in resected stage I non-small-cell lung cancer. BMC Cancer 2020;20:150. [Crossref] [PubMed]
- Yousaf-Khan U, van der Aalst C, de Jong PA, et al. Final screening round of the NELSON lung cancer screening trial: the effect of a 2.5-year screening interval. Thorax 2017;72:48-56. [Crossref] [PubMed]
- Tartarone A, Lerose R, Aieta M. Focus on lung cancer screening. J Thorac Dis 2020;12:3815-20. [Crossref] [PubMed]
- Friedlaender A, Addeo A, Russo A, et al. Targeted Therapies in Early Stage NSCLC: Hype or Hope? Int J Mol Sci 2020;21:6329. [Crossref] [PubMed]
- Hofman P, Rouleau E, Sabourin JC, et al. Predictive molecular pathology in non-small cell lung cancer in France: The past, the present and the perspectives. Cancer Cytopathol 2020;128:601-10. [Crossref] [PubMed]
- VanderLaan PA, Roy-Chowdhuri S. Current and future trends in non-small cell lung cancer biomarker testing: The American experience. Cancer Cytopathol 2020;128:629-36. [Crossref] [PubMed]
- Vishnoi A, Rani S. MiRNA Biogenesis and Regulation of Diseases: An Overview. Methods Mol Biol 2017;1509:1-10. [Crossref] [PubMed]
- Iqbal MA, Arora S, Prakasam G, et al. MicroRNA in lung cancer: role, mechanisms, pathways and therapeutic relevance. Mol Aspects Med 2019;70:3-20. [Crossref] [PubMed]
- Eggert JA, Palavanzadeh M, Blanton A. Screening and Early Detection of Lung Cancer. Semin Oncol Nurs 2017;33:129-40. [Crossref] [PubMed]
- Ghadjar P, Rubie C, Aebersold DM, et al. The chemokine CCL20 and its receptor CCR6 in human malignancy with focus on colorectal cancer. Int J Cancer 2009;125:741-5. [Crossref] [PubMed]
- Zhang XP, Hu ZJ, Meng AH, et al. Role of CCL20/CCR6 and the ERK signaling pathway in lung adenocarcinoma. Oncol Lett 2017;14:8183-9. [Crossref] [PubMed]
- Barrett T, Edgar R. Gene expression omnibus: microarray data storage, submission, retrieval, and analysis. Methods Enzymol 2006;411:352-69. [Crossref] [PubMed]
- Zhao J, Li L, Wang Q, et al. CircRNA Expression Profile in Early-Stage Lung Adenocarcinoma Patients. Cell Physiol Biochem 2017;44:2138-46. [Crossref] [PubMed]
- Jiang N, Zou C, Zhu Y, et al. HIF-1ɑ-regulated miR-1275 maintains stem cell-like phenotypes and promotes the progression of LUAD by simultaneously activating Wnt/β-catenin and Notch signaling. Theranostics 2020;10:2553-70. [Crossref] [PubMed]
- Ma CX, Ellis MJ. The Cancer Genome Atlas: clinical applications for breast cancer. Oncology (Williston Park) 2013;27:1263-9, 1274-9. [PubMed]
- Carithers LJ, Moore HM. The Genotype-Tissue Expression (GTEx) Project. Biopreserv Biobank 2015;13:307-8. [Crossref] [PubMed]
- Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer 2018;17:79. [Crossref] [PubMed]
- Gan TQ, Xie ZC, Tang RX, et al. Clinical value of miR-145-5p in NSCLC and potential molecular mechanism exploration: A retrospective study based on GEO, qRT-PCR, and TCGA data. Tumour Biol 2017;39:1010428317691683. [Crossref] [PubMed]
- Chen L-L, Yang L. Regulation of circRNA biogenesis. RNA Biology 2015;12:381-8. [Crossref] [PubMed]
- Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol 2016;238:42-51. [Crossref] [PubMed]
- Du J, Xu J, Chen J, et al. circRAE1 promotes colorectal cancer cell migration and invasion by modulating miR-338-3p/TYRO3 axis. Cancer Cell Int 2020;20:430. [Crossref] [PubMed]
- Lai Z, Wei T, Li Q, et al. Exosomal circFBLIM1 Promotes Hepatocellular Carcinoma Progression and Glycolysis by Regulating the miR-338/LRP6 Axis. Cancer Biother Radiopharm 2020. Epub ahead of print. [Crossref] [PubMed]
- Lv YS, Wang C, Li LX, et al. Effects of circRNA_103993 on the proliferation and apoptosis of NSCLC cells through miR-1271/ERG signaling pathway. Eur Rev Med Pharmacol Sci 2020;24:8384-93. [PubMed]
- Nanishi K, Konishi H, Shoda K, et al. Circulating circERBB2 as a potential prognostic biomarker for gastric cancer: An investigative study. Cancer Sci 2020;111:4177-86. [Crossref] [PubMed]
- Yin G, Zheng S, Shan Y, et al. Circulating circRNA Has_Circ_0141633 Serves as a Potential Biomarker for Gastric Cancer. Clin Lab 2020. [Crossref] [PubMed]
- Lin Y, Su W, Lan G. Value of circular RNA 0007385 in disease monitoring and prognosis estimation in non-small-cell lung cancer patients. J Clin Lab Anal 2020;34:e23338. [Crossref] [PubMed]
- Luo YH, Yang YP, Chien CS, et al. Plasma Level of Circular RNA hsa_circ_0000190 Correlates with Tumor Progression and Poor Treatment Response in Advanced Lung Cancers. Cancers (Basel) 2020;12:1740. [Crossref] [PubMed]
- Chen F, Huang C, Wu Q, et al. Circular RNAs expression profiles in plasma exosomes from early-stage lung adenocarcinoma and the potential biomarkers. J Cell Biochem 2020;121:2525-33. [Crossref] [PubMed]
- Xian J, Su W, Liu L, et al. Identification of Three Circular RNA Cargoes in Serum Exosomes as Diagnostic Biomarkers of Non-Small-Cell Lung Cancer in the Chinese Population. J Mol Diagn 2020;22:1096-108. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215-33. [Crossref] [PubMed]
- Murphy PM, Baggiolini M, Charo IF, et al. International union of pharmacology. XXII. Nomenclature for chemokine receptors. Pharmacol Rev 2000;52:145-76. [PubMed]
- Schutyser E, Struyf S, Van Damme J. The CC chemokine CCL20 and its receptor CCR6. Cytokine Growth Factor Rev 2003;14:409-26. [Crossref] [PubMed]
- Mantovani A. Chemokines in neoplastic progression. Semin Cancer Biol 2004;14:147-8. [Crossref] [PubMed]
- Zhang XP, Hu ZJ, Meng AH, et al. Role of CCL20/CCR6 and the ERK signaling pathway in lung adenocarcinoma. Oncol Lett 2017;14:8183-9. [Crossref] [PubMed]
- Wang B, Shi L, Sun X, et al. Production of CCL20 from lung cancer cells induces the cell migration and proliferation through PI3K pathway. J Cell Mol Med 2016;20:920-9. [Crossref] [PubMed]
(English Language Editor: J. Teoh)


