
Long non-coding RNA MT1DP interacts with miR-365 and induces apoptosis of nucleus pulposus cells by repressing NRF-2-induced anti-oxidation in lumbar disc herniation
Introduction
Intervertebral disc (IVD) degeneration (IDD) is a common and frequently-occurring disease in clinical practice that can cause a series of clinical syndromes, including cervical spondylosis and disc herniation (1). Current research suggests that IDD occurs under a variety of physiological and pathological conditions that can be affected by a range of factors, such as genetics, cellular senescence, mechanical load, matrix degradation, inflammation, and apoptosis. The decrease in the number of nucleus pulposus cells (NPCs) and the loss of extracellular matrix (ECM) are the core characteristics of IDD (2). However, there is no unified theory on the exact mechanisms of IDD. Once IDD occurs, it is difficult to reverse, and there is currently no effective means to deter its progression (3). Therefore, it is necessary to conduct in depth basic research to understand the mechanisms of IDD.
Long noncoding RNA (lncRNA) is a group of endogenous, non-coding transcripts of more than 200 nucleotides in length. Increasingly, evidence suggests that lncRNAs act as key regulators in biological processes. Abnormal expression of lncRNAs has been shown to be involved in a variety of human diseases, including IDD and osteoarthritis (4). However, understanding the mechanisms by which lncRNA regulates IDD is still in its infancy.
As an oncogene, MT1DP has been shown to be involved in cell proliferation, apoptosis, differentiation and metastasis (5). Recently, Gao et al. (6) demonstrated that MT1DP induces apoptosis by suppressing the anti-oxidant activity of the nuclear factor erythroid 2-related factor 2 (NRF-2). The interaction between the microRNA (miRNA) miR-365 and NRF-2 was shown to inhibit oxidative stress-induced apoptosis in cancer cells (7). Interestingly, NRF-2 expression has been shown to be correlated with IDD (8). However, whether MT1DP plays a role in the pathogenesis of IDD and the mechanisms involved remain to be fully elucidated. It has been suggested that lncRNAs can competitively adsorb certain miRNAs through endogenous RNA pathways to regulate gene expression and thereby participate in the development of IDD (9). Gao et al. (6) confirmed that the lncRNA MT1DP acts as an endogenous sponge to inhibit cell proliferation and induce apoptosis by directly binding to miR-365 and this may promote the development of IDD. We previously found out that miR-365 is a potential target of MT1DP through data base analysis. We speculated that MT1DP may participate in the development of IDD by targeting miR-365 to regulate NRF-2. This investigation used human degenerative IVD NPCs in vitro to investigate the roles and mechanisms of MT1DP and NRF-2 in IDD. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-8123).
Methods
Patients and samples
Human degenerative IVD nuclear pulposus (NP) tissues were obtained from 10 patients with lumbar disc herniation (LDH) who underwent lumbar spine surgery (the IDD group). This group included 5 males and 5 females, aged 45–65 years (mean age 52±10 years). All specimens were collected between March 2017 and March 2019 in the Orthopedic Department of The Second Affiliated Hospital of Guangzhou Medical University. LDH was diagnosed by preoperative symptoms, clinical signs, and imaging. Patients in whom conservative treatment was ineffective subsequently underwent surgery. Postoperative pathological findings in these patients confirmed degenerative disc tissues, with grades of III–IV according to the Pfirrmann degenerative IVD sagittal MRI (magnetic resonance imaging) grading standard (10).
Normal IVD NP tissues were obtained from 10 patients with lumbar vertebrae fractures. This group was referred to as the negative control (NC) group and included 5 males and 5 females (aged 45–65 years, mean age 53±11 years). Intraoperative human NP tissues were placed in Dulbecco’s modified Eagle’s medium (DMEM)/F12 medium (HyClone, USA) containing 10% fetal bovine serum (FBS; Gibco, USA), and stored at 4 °C prior to subsequent processing. This study was approved by the Ethics Committee of The Second Affiliated Hospital of Guangzhou Medical University (No. B2020-052). This study conformed to the provisions of the Declaration of Helsinki (as revised in 2013). All participants gave informed consent.
Isolation, culture, and identification of the NPCs
The NP tissues were dissected with ophthalmic scissors into 1 mm3 pieces and incubated with 0.25% trypsin for 30 minutes followed by centrifugation at 1,000 rpm for 10 minutes. The supernatant was discarded, and the tissue was digested with type II collagenase was for 4 hours at 37 °C. After filtration through a 200-mesh filter and centrifugation, the cells were collected. The NPCs were resuspended in DMEM/F12 medium (ThermoFisher, USA) containing 20% FBS (Biosera, USA) and cultured in a 37 °C, 5% CO2 incubator. The cells were cultured to 70–80% confluence, trypsinized, and passaged at 1:2. The second passage of the NPCs were seeded into 6-well plates for preparation of slides. NPCs were identified by immunocytochemical staining of collagen II (11) and Safranin O staining (12) as described previously.
Cell transfection
The MT1DP sequence was synthesized by Sangon Biotech (Shanghai, China) and cloned into the pcDNA3.1 plasmid (Thermo Fisher Scientific, USA) to construct the MT1DP overexpression vector (the MT1DP group). The pcDNA3.1 empty vector was used as a NC (the NC group). The pcDNA-miR-365 and the corresponding control pcDNA-Ctrl were also synthesized and constructed by Sangon Biotech. Plasmids or oligonucleotides were transfected into passage 3 degenerative IVD NPCs using a Lipofectamine 3000 Transfection kit (Life Technologies, USA) according to the manufacturer’s instructions.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA in the NP tissues and the NPCs were extracted using the Total RNA Rapid Extraction Kit (Sigma-Aldrich, USA). Total RNA (500 ng) was used to analyze the relative expressions of NRF-2, MT1DP, and miR-365 by using the SYBR Green PCR kit (Takara, Japan) on a 7500 fast real-time PCR system (Applied Biosystems, USA). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and U6 were used as internal parameters. The relative expression of the gene of interest was calculated by the 2-ΔΔt method.
The primers used were as follows: NRF2: 5'-C ACATCCAGTCAGAAACCAGTGG-3' and 5'-GGAAT GTCTGCGCCAAAAGCTG-3'; MT1DP: 5'-TCAAGG CCAAAGGTGGCTCCTGCAC-3', and 5'-GCACGGCA GCTGCACTTCACCAATG-3'; miR-365a-3p: 5'-TGCG GTAATGCCCCTAAAAA-3' and 5'-TGCAAGAGCAAT AAGGATT-3'; U6: 5'-CTCGCTTCGGCAGCACA-3' and 5'-AACGCTTCACGAATTTGCGT-3'; and GAPDH: 5'-CAATGACCCCTTCATTGACC-3' and 5'-GACAAG CTTCCCGTTCTCAG-3'.
Cell counting Kit 8 (CCK-8) assay
Cells (1×103 cells/well) were seeded in a 96-well plate and treated with hydrogen peroxide (H2O2), NRF-2 inhibitor, or MT1DP + miR-365 over-expression for 48 hours. CCK-8 (Beyotime, China) solution (1:10) was added to the cells and incubated for 4 hours. The absorbance was measured at 490 nm by a SpectraMax M5 plate reader (Molecular Devices, Sunnyvale, CA, USA).
Western blotting analysis
Cells were completely lysed in lysis buffer (Takara, Japan) and nuclear proteins and cytoplasmic proteins were extracted. Protein concentrations were determined using the BCA Protein Assay Kit (Pierce). SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis) was performed with 50 µg of the sample, and transferred onto PVDF (polyvinylidene difluoride) membranes. Membranes were then blocked with 5% bovine serum albumin (BSA) for 1 hour at room temperature and subsequently incubated with the corresponding primary antibodies, including anti-Bax (Abcam, USA), anti-B cell lymphoma 2 (Bcl-2; Abcam, USA), anti-cleaved-caspase 3 (Abcam, USA), anti-CYCS (anti-cytochrome C somatic; Abcam, USA), anti-cyclooxygenase 4 (anti-COX4; Abcam, USA), anti-voltage-dependent anion channel 1 (anti-VDAC 1; Abcam, USA) and anti-GAPDH (Abcam, USA) overnight at 4 °C. GAPDH was used as the internal control. After washing with tris-buffered saline with Tween20 (TBST), membranes were incubated with horseradish peroxidase-labeled secondary antibody for 1 hour at room temperature. Following TBST washes, specific protein bands were visualized using an enhanced chemiluminescent solution (ECL; Millipore, Shanghai, Chian).
JC-1 staining
Fluorescent dye 5,5’,6,6’-tetrachloro-1,1’,3,3’-tetraethyl benzimidazolyl-carbocyanineio-dide (JC-1; Beyotime Biotech, Nanjing, China) was used to monitor the mitochondrial membrane potential (ΔψM). Cells were seeded into 6-well plates at 3×105 cells/well.
Reactive oxygen species (ROS) detection
Cells were transferred to a 6-well plate (3×105) and allowed to attach for 6 h at 37 °C. To detect mitochondrial-related ROS accumulation, 0.5 mM H2O2 was added into medium for 24 h and subsequently MitoSOX red (Thermo Fisher Scientific, Inc.) staining was performed. Cells were plated in serum-free supplemented with 0.7 µM MitoSOX for 3 min, and the fluorescent signal at a wavelength of 525 nm was detected using a Lumina Fluorescence Spectrometer (Thermo Fisher Scientific, Inc.)
Immunofluorescence staining
The NP cells were seeded onto cover slips in 24-well plates. After the corresponding treatments, the slides with the NP cells were washed with pre-cooled phosphate buffered saline (PBS). Cells were fixed for 10 minutes in 200 µL of 4% paraformaldehyde per well. After washing, the cells were incubated with 0.1% Triton X-100 for 10 minutes at room temperature, followed by 5% goat serum for 30 minutes. Cells were then incubated with 200 µL of reactive oxygen species (ROS) probe (Beyotime, China) for 1 hour at room temperature. After washing, the slides were removed from the 24-well plates and a small amount of anti-fluorescence quenching was added. The images were observed and captured under a Carl Zeiss LSM700 laser scanning confocal microscope (Zeiss, Germany).
Statistical analysis
Statistical analyses were performed by Student’s t-test or one-way analysis of variance (ANOVA) using software SPSS 15.0 (SPSS Inc., Chicago, IL, USA). All data were presented as the mean ± standard deviation (mean ± SD).
Results
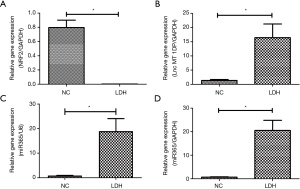
The expression of NRF2 decreased, while MT1DP and miR-365 increased in patients with herniated disc
The expression of NRF-2 in LDH (as the IDD group) was significantly down-regulated compared to the NC group (P<0.05; Figure 1A). Meanwhile the expression of MT1DP and miR-365 in the LDH group was significantly up-regulated compared to the NC group (P<0.05; Figure 1B,C). The interaction between MT1DP and miR-365 was examined. The results of the RNA immunoprecipitation (RIP) assay revealed a direct interaction between MT1DP and miR-365 (P<0.05; Figure 1D).
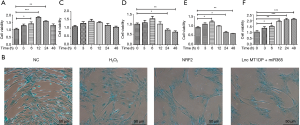
MT1DP and miR-365 affected NP cell viability in a time-dependent manner
The viability of the NP cells was assessed over a 48-hour period. Increased viability was observed in the first 12 hours and this decreased during the next 12 hours (significant difference P value: *P<0.05, **P<0.01, ***P<0.0001; Figure 2A,B). Oxidative damage by H2O2 neutralized any change of viability in the cell cultures and no significant changes in cell viability was detected (Figure 2C). The specific NRF-2 inhibitor ML385 not only suppressed the viability of the NP cells but it also caused cytotoxicity. A significant decline in cell viability was noted by 24 hours and this declined further by 48 hours (*P<0.05, **P<001; Figure 2D). Interestingly, the same trend was observed in cells over-expressing MT1DP + miR-365. The increased viability of the NP cells was suppressed and this further declined by 24 and 48 hours (*P<0.05, **P<0.01; Figure 2E). Similar results were observed by cell morphology and density analyses using Allison Blue staining. Fewer cells were observed in both the NRF-2 inhibition group and the MT1DP + miR-365 over-expression group (Figure 2F).
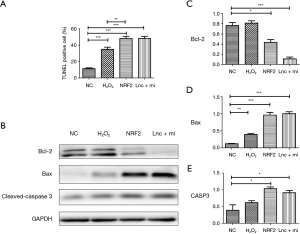
Lnc-MT1DP + miR-365 inhibited NRF-2-induced apoptosis of NP cells
Treatment of NP cells with H2O2 resulted in cell apoptosis. Both inhibition of NFR-2 and overexpression of MT1DP + miR-365 enhanced the oxidative-induced apoptosis (*P<0.05, **P<0.01, ***P<0.0001; Figure 3A). Simultaneously, the pro-apoptosis protein Bax and Caspase 3 expression were elevated while the anti-apoptosis protein Bcl-2 was suppressed in inhibition of NRF-2 or overexpression of MT1DP + miR-365 NPCs (*P<0.05, **P<0.01, ***P<0.0001; Figure 3B,C,D,E), suggesting that alterations in these proteins play a role in the oxidative damage-induced apoptosis of NP cells.
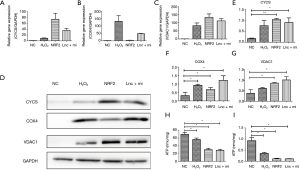
Lnc-MT1DP + miR-365 caused inflammation in NP cells by damaging the mitochondrial membrane
The effects of MT1DP + miR-365 on the mitochondria were investigated. In cells damaged by H2O2-induced oxidative stress, mitochondrial damage was assessed by evaluating the expression of CYCS, COX4, and VDAC1. The mRNA levels of CYCS, COX4, and VDAC1 were measured by qRT-PCR. The results revealed up-regulation of CYCS, COX4, and VDAC1 (*P<0.05; Figure 4A,B,C). The expressions of these three proteins were significantly increased by NRF-2 inhibition and MT1DP + miR-365 over-expression compared to control cells, indicating that NRF-2 inhibition and MT1DP + miR-365 over-expression caused further oxidative damage in the mitochondria (*P<0.05, **P<0.01; Figure 4D,E,F,G). The relative expression of ATP and mtDNA are significantly reduced by NRF-2 inhibition and MT1DP + miR-365 over-expression compared to control cells, indicating that NRF-2 inhibition and MT1DP + miR-365 over-expression caused the mitochondria dysfunction (*P<0.05; Figure 4H,I).
The combination of lnc-MT1DP and miR-365 reduced cell mitochondrial function and decreased the ability of cells to eliminate ROS
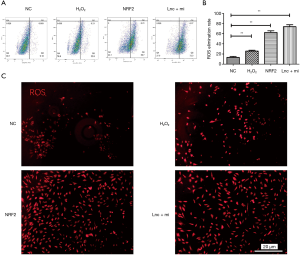
Flow cytometry analyses confirmed that overexpression of MT1DP + miR365 induced lower levels of mitochondrial membrane potential and higher levels of ROS compared to NC cells (**P<0.01, Figure 5A,B). Meanwhile, ROS fluorescence was analyzed to explore the biological effects of NRF-2 inhibition and MT1DP + miR365 over-expression on NP cells. As expected, the ROS levels following NRF-2 inhibition and MT1DP + miR365 over-expression increased significantly (Figure 5C).
Discussion
Degeneration of the IVD is accompanied by a decrease in the number of cells and reduced synthesis of the ECM (13). Recently, the role of lncRNAs in IDD has received extensive attention. In this report, we demonstrated that MT1DP was significantly up-regulated in human degenerative disc NP tissue. Additionally, MT1DP participated in the promotion of IDD by inhibiting cell proliferation, and promoting apoptosis and ECM degradation of human degenerative disc NP cells in vitro. More importantly, MT1DP promoted IDD by targeting miR-365 to suppress the NRF-2 antioxidant signaling pathway.
Recently, some studies have reported significant changes in the expression of many lncRNAs in IDD (4). Chen et al. (9) reported that 8 lncRNAs were differentially expressed in human degenerative NPCs compared to normal NPCs. Wan et al. (14) also found that 116 lncRNAs were differentially expressed in IDD, and overexpression of the lncRNA RP11 296A18.3 induced up-regulation of Fas-associated protein factor 1 (FAF1) and ultimately enhanced abnormal apoptosis of IVD NPCs. Exploring the mechanisms by which lncRNA regulate the development of IDD may have important implications for the discovery of new therapeutic targets. MT1DP, a type of lncRNA, has been shown to be highly expressed in a variety of malignancies and is closely related to the development of tumors (5,7,15). Recent studies have found that MT1DP is closely related to the miR-365/NRF-2 axis (6). NRF-2 has been shown to play a critical role in IDD development via various pathways including the AKT-ERK1/2-NRF-2 signaling pathway (16), the Keap1/NRF-2/p62 feedback loop (8), and the NRF-2/ARE and MARK signaling pathways (17). All these studies indicated that NRF-2 has a central function in IDD development, which suggests that MT1DP/miR-365 may also have some regulatory effect in the development of IDD. The present study also demonstrated that MT1DP expression was significantly up-regulated in degenerated IVD NP tissues and cells, suggesting that MT1DP may be involved in the occurrence and development of IDD.
Apoptosis of the NPCs of the IVD plays an important role in the IDD process, of which the mitochondrial apoptosis pathway has also been shown to be important (18). Mitochondria regulate endogenous and exogenous apoptotic pathways through caspase-dependent and non-caspase-dependent pathways. Bcl-2 and caspases are evolutionarily conserved protein families in eukaryotes. Bcl-2 can prevent the release of cytochrome C from the mitochondria to the cytoplasm thereby inhibiting the apoptosis of NPCs, while caspases mediate the initiation and execution of apoptosis. Caspase-3 is the caspase involved in apoptosis, and silencing caspase-3 in vitro by mechanical overload can reduce the apoptosis of NPCs, thereby inhibiting IDD.(19) This current study demonstrated that the expression of these two key protein family members was altered following MT1DP overexpression in NPCs, suggesting that MP1DT could activate the mitochondrial apoptosis pathway in NPCs by down-regulating Bcl-2 and up-regulating caspase-3. These results, together with reports in the literature, suggested that MT1DP regulated the balance between pro-apoptotic and apoptotic molecules in the mitochondrial apoptotic pathway, and the expression of MP1DP determined the sensitivity of NPCs to apoptotic stimuli.
Limit to the sample sizes, we had to admit there's insufficiency in sample imaging but we'll further expand the sample of each group and display some more convincing proofs.
Conclusions
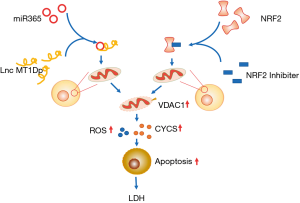
The combination of lncRNA MT1DP and miR-365 damaged the cell mitochondrial membrane, reduced mitochondrial function and the ability to eliminate ROS, and increased cell apoptosis, thereby leading to LDH. MT1DP played a role in the promotion of apoptosis of the degenerated IVD NPCs by targeting miR-365, which may be related to the inhibition of the NRF-2 pathway. Our study further elucidated the mechanisms by which MT1DP promotes the progression of IDD, and this may provide novel ideas for the treatment of degenerative diseases (Figure 6).
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of Guangdong (Grant No. 2019A1515010690).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-8123
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-8123
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-8123). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethics Committee of The Second Affiliated Hospital of Guangzhou Medical University (No. B2020-052). This study conformed to the provisions of the Declaration of Helsinki (as revised in 2013). All participants gave informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- van Uden S, Silva-Correia J, Oliveira JM, et al. Current strategies for treatment of intervertebral disc degeneration: substitution and regeneration possibilities. Biomater Res 2017;21:22. [Crossref] [PubMed]
- Cappello R, Bird JLE, Pfeiffer D, et al. Notochordal Cell Produce and Assemble Extracellular Matrix in a Distinct Manner, Which May Be Responsible for the Maintenance of Healthy Nucleus Pulposus. Spine (Phila Pa 1976) 2006;31:873-82; discussion 883. [Crossref] [PubMed]
- Sakai D, Schol J. Cell therapy for intervertebral disc repair: Clinical perspective. J Orthop Translat 2017;9:8-18. [Crossref] [PubMed]
- Chen W, Yu X, Yang W, et al. lncRNAs: novel players in intervertebral disc degeneration and osteoarthritis. Cell Prolif 2017;50:e12313. [Crossref] [PubMed]
- Yu W, Qiao Y, Tang X, et al. Tumor suppressor long non-coding RNA, MT1DP is negatively regulated by YAP and Runx2 to inhibit FoxA1 in liver cancer cells. Cell Signal 2014;26:2961-8. [Crossref] [PubMed]
- Gao M, Li C, Xu M, et al. LncRNA MT1DP Aggravates Cadmium-Induced Oxidative Stress by Repressing the Function of Nrf2 and is Dependent on Interaction with miR-365. Adv Sci (Weinh) 2018;5:1800087. [Crossref] [PubMed]
- Gai C, Liu C, Wu X, et al. MT1DP loaded by folate-modified liposomes sensitizes erastin-induced ferroptosis via regulating miR-365a-3p/NRF2 axis in non-small cell lung cancer cells. Cell Death Dis 2020;11:751. [Crossref] [PubMed]
- Tang Z, Hu B, Zang F, et al. Nrf2 drives oxidative stress-induced autophagy in nucleus pulposus cells via a Keap1/Nrf2/p62 feedback loop to protect intervertebral disc from degeneration. Cell Death Dis 2019;10:510. [Crossref] [PubMed]
- Chen Y, Ni H, Zhao Y, et al. Potential Role of lncRNAs in Contributing to Pathogenesis of Intervertebral Disc Degeneration Based on Microarray Data. Med Sci Monit 2015;21:3449-58. [Crossref] [PubMed]
- Pfirrmann CWA, Metzdorf A, Zanetti M, et al. Magnetic Resonance Classification of Lumbar Intervertebral Disc Degeneration. Spine (Phila Pa 1976) 2001;26:1873-78. [Crossref] [PubMed]
- Song X, Peng S. Transplantation of transforming growth factor beta3 gene-modified nucleus pulposus cells for intervertebral disc degeneration in rabbits. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi 2012;26:790-5. [PubMed]
- Wang D, Vo NV, Sowa GA, et al. Bupivacaine decreases cell viability and matrix protein synthesis in an intervertebral disc organ model system. Spine J 2011;11:139-46. [Crossref] [PubMed]
- Adams MA, Roughley PJ. What is intervertebral disc degeneration, and what causes it? Spine (Phila Pa 1976) 2006;31:2151-61. [Crossref] [PubMed]
- Wan ZY, Song F, Sun Z, et al. Aberrantly expressed long noncoding RNAs in human intervertebral disc degeneration: a microarray related study. Arthritis Res Ther 2014;16:465. [Crossref] [PubMed]
- Gao M, Chen M, Li C, et al. Long non-coding RNA MT1DP shunts the cellular defense to cytotoxicity through crosstalk with MT1H and RhoC in cadmium stress. Cell Discov 2018;4:5-19. [Crossref] [PubMed]
- Wang Y, Zuo R, Wang Z, et al. Kinsenoside ameliorates intervertebral disc degeneration through the activation of AKT-ERK1/2-Nrf2 signaling pathway. Aging (Albany NY) 2019;11:7961-77. [Crossref] [PubMed]
- Fang W, Zhou X, Wang J, et al. Wogonin mitigates intervertebral disc degeneration through the Nrf2/ARE and MAPK signaling pathways. Int Immunopharmacol 2018;65:539-49. [Crossref] [PubMed]
- Shi L, Teng H, Zhu M, et al. Paeoniflorin inhibits nucleus pulposus cell apoptosis by regulating the expression of Bcl-2 family proteins and caspase-9 in a rabbit model of intervertebral disc degeneration. Exp Ther Med 2015;10:257-62. [Crossref] [PubMed]
- Yamada K, Sudo H, Iwasaki K, et al. Caspase 3 silencing inhibits biomechanical overload-induced intervertebral disk degeneration. Am J Pathol 2014;184:753-64. [Crossref] [PubMed]
(English Language Editor: J. Teoh)


