
Development and validation of a novel nomogram to predict cancer-specific survival in patients with uterine cervical adenocarcinoma
Introduction
Uterine cervical cancer has shown a steady decline in overall incidence and mortality in the United States; however, it has remained a serious health threat in developing countries (1,2). Uterine cervical adenocarcinoma (UAC), the second most common histologic type of invasive cervical carcinoma, has shown an upward trend in both true and relative incidence over the past two decades (3).
According to the National Comprehensive Cancer Network (NCCN) guidelines, the current treatment strategies and prognostic factors for squamous cell carcinoma (SCC) were employed for UAC, which is primarily determined by the International Federation of Gynecology and Obstetrics (FIGO) stage (4). Although retroperitoneal lymph node status was incorporated into the revised FIGO staging system in 2018, FIGO stage is still a clinically based staging system (5), and patients with the same FIGO stage often have different prognoses, especially those with large tumor size or endocervical adenocarcinoma. Increasing evidence has suggested that the biological behavior, treatment outcomes, and prognostic factors of UAC differ from those of SCC. Hu et al. (6) demonstrated that patients with UAC had poorer overall survival (OS) and disease-free survival (DFS) than those with stage IB–IVA SCC, regardless of whether they were treated with radiotherapy alone or concurrent chemoradiotherapy (CCRT). Therefore, it is necessary to develop an individualized predictive model that supplements the FIGO staging system for predicting prognosis and optimizing treatment strategies in UAC. However, due to its relatively low incidence, few studies on UAC have focused on the prediction of patient prognosis or treatment outcomes (7-9).
Prediction models have been widely accepted as reliable tools to aid in the assessment of prognosis and clinical decision-making for numerous malignancies, including uterine SCC, prostate cancer, pulmonary carcinoma, and gastric cancer (10-13). In this study based on the Surveillance, Epidemiology, and End Results (SEER) database, the prognostic factors predicting cancer-specific death in UAC patients were explored, and a practical prognostic model was constructed and validated for optimal clinical management and individualized patient counselling. We present the following article in accordance with the transparent reporting of a multivariable prediction model for prognosis or diagnosis (TRIPOD) reporting checklist (available at: http://dx.doi.org/10.21037/atm-20-6201).
Methods
Study design and data extraction
Data from the SEER database (SEER 18 registries, November 2019 data submission), which covers approximately 27.8% of the U.S. population, were extracted using SEER*Stat 8.3.6 software (www.seer.cancer.gov) and retrospectively analyzed (14,15). Cases with malignant adenocarcinomas were identified using the International Classification of Diseases for Oncology, third edition (ICD-O-3). The primary tumor site codes were: C53.0-53.1 and C53.8-53.9 (uterine cervix). The histology codes were: 8140/3, 8144/3, 8147/3, 8200/3, 8210/3, 8241/3, 8244/3, 8255/3, 8260/3, 8261/3, 8262/3, 8263/3, 8310/3, 8313/3, 8323/3, 8380/3, 8382/3, 8384/3, 8430/3, 8441/3, 8460/3, 8461/3, 8480/3, 8481/3, 8482/3, and 8490/3 (16,17).
The inclusion criteria were: histologically proven malignant adenocarcinoma as the first primary tumor; diagnosed between 1988 and 2016; and known survival months and cause of death. The exclusion criteria were: diagnosed with cervical intraepithelial neoplasia (CIN) or another histologic type of cervical cancer including SCC, undifferentiated carcinoma, neuroendocrine tumors, adeno-squamous carcinoma, mesenchymal tumors, or mixed epithelial and mesenchymal tumors as adenosarcoma; diagnosed through autopsy or death certificate; unknown cause of death; and follow-up time or survival time unavailable.
Institutional review board approval was not required due to the SEER database being an open public database. Signed authorization and received permission were provided by the SEER program for access and use of the dataset.
Since most data of the participants diagnosed between 1973 and 1988 were unavailable, January 1988 was set as the starting point of the study. Patients diagnosed between 1988 and 2010 (n=5,655) were used to develop the model, and those diagnosed between 2011 and 2016 (n=3,336) were used for temporal validation. The process flowchart is presented in Figure 1. The following data were collected from the SEER database: age, race, year of diagnosis, histologic subtype, grade, SEER summary stage (localized, regional, or distant), American Joint Committee on Cancer (AJCC) staging system tumor, node, metastasis (TNM) stage, AJCC stage, surgery of the primary site, surgery of regional lymph nodes (LNs), surgery of metastasis, radiation, chemotherapy, tumor size, number of regional LNs examined, number of positive LNs, sequence number of multi-primary tumors, survival time, cause of death, and vital status.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) (18).
Outcomes
The primary endpoint of the study was death specifically attributed to UAC. The survival time was the interval from the date of diagnosis to the date of death or the last follow-up. Participants with an unknown cause of death, unknown survival time, or a survival time of zero were excluded from the study. Participants who were alive at the time of last follow-up were counted as censored observations.
Predictors
The following variables acquired from the SEER database were incorporated into further analyses: age, race, grade, AJCC TNM stage, surgery of the primary site (yes or no), surgery of regional lymph nodes (LNs) (yes or no), surgery of metastasis (yes or no), radiation (yes or no), chemotherapy (yes or no), tumor size, number of regional LNs examined, number of positive LNs, and sequence number of multi-primary tumors. These 15 factors were included in the subsequent analysis as candidate predictors, which determined the minimum number of outcome events and sample size of the study.
Handling of missing data
The percentage of missing values in the present study was <20%, which were as follows: 52 (0.5%) cases with race missing, 1,596 (17.7%) cases with grade missing, 233 (2.5%) cases with number of LNs examined missing, and 816 (9.0%) cases with the number of positive LNs missing. To control bias and reach valid conclusions for the target cohort, we analyzed the correlation between the variables with missing data and assumed the data were missing at random. The missing data were handled with a multiple imputation method by chained equations for a total of 20 times (19). In all, 2,697 (1.4%) of the 188,811 values were multi-imputed for further analysis.
Transformation of the predictors
After analysis with restricted cubic splines, the associations between continuous variables and outcomes were found to be nonlinearly related (data not shown). The continuous variables were further confirmed to have a non-normal distribution, even after log-transformation. Hence, they were converted into categorical variables. Age at diagnosis was categorized as: ≤39, 40–59, >60 years. Tumor size, defined as the largest measurement of horizontal spread or surface diameter in the field, was stratified into three groups according to the FIGO staging system: ≤20, 21–40, and >40 mm (5). Based on the U-shaped relation between the number of dissected LNs and cause-specific mortality (data not shown), the number of LNs examined was classified as: 0, 1–15, 16–30, and >30. The number of positive LNs was grouped as: 0, 1–3, and >3, according to previous studies (20). With regard to tumor grade, cases with grade III and IV were incorporated as a single group due to the limited number of grade IV (270, 3%) cases. Thus, tumor grade was reclassified into three groups: grade I, grade II, and grade III plus IV. SEER summary stage was categorized into localized, regional, and distant groupings. The regional group included those with direct extension only, those with both direct extension and lymph node involvement, and those with lymph node involvement only. The distant group included the cases with distant site/nodal involvement. The AJCC stage and AJCC stage T were reclassified as IA, IB, II, III, IV, and T1a, T1b, T2, T3, and T4, respectively. Due to the difference in prognosis between IA and IB. AJCC stage N and stage M were described as N0, N1, and M0, M1. Rather than having 3 groups for the sequence number of multi-primary tumors, 2 groups (the 1 primary only group and the 1st of 2 or more primaries group) were formed, because 700 patients with unknown causes of death were excluded and for them, UAC was found as the second or more of primaries.
Predictor selection
To identify the potential risk factors for cancer-specific death (CSD), all the predictors were enrolled into a least absolute shrinkage and selection operator (LASSO) regression analysis with the development cohort. The LASSO regression analysis was used via cross-validation to prevent overfitting of the variables from the development dataset, which penalized the absolute value of the regression coefficients. With larger penalties, the estimates of weaker factors shrank toward zero, so that only the most effective predictors were retained in the model. LASSO regression analysis was applied to determine the most important predictors from the derivation dataset. In the process, a total of 15 variables with non-zero coefficient value were identified with corresponding lambda value and likelihood of deviance. Ten variables were included with 1 standard error criterion (Figure S1). To refine the model for clinical use, the predictive power of the models with 5 to 10 variables were compared. The results showed the area under the ROC (AUC) varied from 0.848 to 0.853 when 7 to 10 variables were incorporated into the model, whereas AUC of the model was 0.837 with 6 variables (data not shown). Therefore, 7 variables with the corresponding lambda value were ascertained into subsequent analysis: grade, stage T, stage N, stage M, tumor size, number of positive LNs and surgery of the primary site. In the multi-collinearity analysis performed among the 7 variables, the variance inflation factor (VIF) was 5.16 and 5.22 for stage N and number of positive LNs respectively, whereas VIFs for the other 5 variables were all <2 (data not shown). This result revealed the multi-collinearity between the two variables, which would have reduced the stability and accuracy of the model. Therefore, the number of positive LNs was excluded. Consequently, 6 variables, including grade, stage T, stage N, stage M, size of the tumor, and surgery of the primary site, were recruited into the final model.
Model development and validation
The model was developed using the Cox proportional hazards risk regression analysis. The Cox proportional hazards assumption was tested and proved to be satisfied for the validity of the estimators (data not shown). To visualize and quantify the effect of each selected variable on the estimated 3- and 5-year cancer-specific survival (CSS) probability, a nomogram was further constructed and validated. A bootstrap resampling method was used for internal validation, in which replacement from the development cohort was performed and the Cox regression models were fitted in 500 bootstrapping replicates. Receiver operating characteristic (ROC) curves and calibration curves for 3- and 5-year CSS were depicted separately. Discrimination could be quantified with AUC or concordance (c)-statistic. Three optimism-adjusted statistics—Harrell’s c-statistic, time-dependent c-statistic, and Brier score—were computed. The dataset from the SEER database diagnosed between 2011 and 2016 (n=3,336) was used for external temporal validation, and the performance of the model was further estimated using the c-statistic, Brier score, baseline survival, calibration slope, and calibration curve. To allow clinicians to calculate CSS with 95% confidence intervals (CI), a free-access online calculator was developed based on the dynamic nomogram (https://betteryuan66.shinyapps.io/dynnomapp/). The c-statistic of the nomogram was also compared to those of AJCC stage and SEER summary stage, with a larger c-statistic indicating a higher prognostic prediction accuracy.
Statistical analysis
Continuous variables were described as mean ± standard deviation (SD) or median with interquartile range (IQR) values depending on whether they had normal or non-normal distribution. Categorical variables were displayed with numbers and percentages per group. LASSO regression analysis was used to select predictors of CSS. Cox hazard regression analysis was used to construct the predictive model, which was presented as a static nomogram and web-based dynamic nomogram. The nomogram was internally validated with a bootstrap resampling method and temporal validation. Using the prediction model, the risk score was calculated for each patient in the development cohort, and they were individually stratified into a low- or high-risk group according to the optimal cutoff value calculated with X-tile software (Yale University, New Haven, CT, USA) (21). Cases with the same cut-off values of risk score in the validation cohort were pooled. Survival curves were depicted using Kaplan-Meier analysis and compared with log-rank test in both the development and validation cohorts. All the statistical analyses were conducted with R software (https://www.r-project.org/, version 3.6.3). The R packages glmnet, MASS, rms, survival, survminer, and DynNom were used for the analysis. A P value <0.05 was considered statistically significant.
Results
Demographic and clinical characteristics
A total of 8,991 patients diagnosed with UAC between 1988 and 2016 from the SEER database were included in our study. The cases diagnosed between 1988 and 2010 (n=5,665) were enrolled for the development of the predictive model, and those diagnosed between 2011 and 2016 (n=3,336) were used for temporal validation. The patients’ baseline characteristics are summarized in Table 1. The entire cohort had a median age of 45, with an IQR of 38–56 years. The 3- and 5-year survival rates (standard error) were 84.0% (0.4) and 79.6% (0.5), respectively, for all patients. Of the patients, 2,739 (31.1%), 4,492 (50.0%), and 1,706 (19.0%) were aged ≤39, 40–59, and ≥60 years, respectively; and 3,934 (43.8%), 2,460 (27.4%), and 2,597 (28.9%) had a tumor size of ≤20, 21–40, and ≥41 mm, respectively. Table S1 presents the characteristics of the development and validation cohorts from after the handling of missing data and reclassification. The number of patients with 0, 1–3, and >3 positive LNs was 7,365 (81.9%), 1,139 (12.7%), and 487 (5.4%), respectively; and the number of patients with 0, 1–15, 16–30, and >30 LNs examined was 3,283 (36.5%), 2,407 (26.8%), 2,467 (27.4%), and 834 (9.3%), respectively.

Full table
Predictor selection
To identify the independent risk factors for CSS, all the candidate variables were enrolled into multivariate Cox regression analysis. As shown in Table 2, 12 of the 15 variables were found to be independently correlated with prognosis for UAC-specific survival (all P<0.05); the exceptions were surgery of regional LNs, surgery of distant metastases, and number of positive LNs. To prevent over-fitting of the variables and to simplify the model, LASSO regression analysis was used to penalize the absolute value of the coefficients (Figure S1). Combined with the results of the LASSO and multi-collinearity analysis, six variables, including grade, stage T, stage N, stage M, size of the tumor, and surgery of the primary site, were entered into the construction of the static nomogram and web-based dynamic nomogram (Table S2).

Full table
Construction of the nomogram
The Cox proportional hazards model was fitted with tumor grade, stage T, stage N, stage M, tumor size, and surgery of the primary site. The results were summarized in a forest plot, which demonstrated the independent impact of the predictors of CSS on UAC patients using hazard ratio (HR) and 95% CI (Figure 2). With regard to tumor grade, using well-differentiated tumor as a reference, moderately differentiated and poorly or undifferentiated tumors were associated with a relatively worse prognosis (HR =1.41, 95% CI: 1.20–1.67, P<0.001; HR =2.10, 95% CI: 1.78–2.48, P<0.001, respectively). In comparison with stage T1a, stage T1b, T2, T3, and T4 were associated with poor prognosis (HR =3.74, 95% CI: 2.45–5.70, P<0.001; HR =7.87, 95% CI: 5.07–12.20, P<0.001; HR =12.81, 95% CI: 8.14–20.14, P<0.001; HR =14.10, 95% CI: 8.77–22.69, P<0.001, respectively). Both the presence of lymph node metastases (HR =1.79, 95% CI: 1.58–2.02, P<0.001) and the presence of distant metastases (HR =1.71, 95% CI: 1.44–2.03, P<0.001) was associated with poor prognosis. The prognosis was poorer with tumor sizes of 21–40 and >40 mm (HR =1.90, 95% CI: 1.58–2.29, P<0.001; HR =2.35, 95% CI: 1.93–2.84, P<0.001, respectively) than with tumor sizes of <21 mm. Surgery of the primary site was identified as an advantageous factor for prognosis (HR =0.56, 95% CI: 0.49–0.63, P<0.001). Based on the model, estimated regression coefficients were converted to point values and the sum of the points was used to predict the probability of 3- and 5-year CSS, which was shown in the nomogram (Figure 3). Detailed points of each prognostic factor in the nomogram are listed in Table S3.


Internal validation and temporal validation of the nomogram
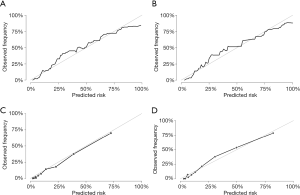
The overall performance of the nomogram was assessed, producing a c-statistic of 0.85 (95% CI: 0.84–0.86). After internal validations with bootstrap resampling, the optimism-adjusted c-statistics at 3 and 5 years were 0.90 (95% CI: 0.89–0.91) and 0.89 (95% CI: 0.88–0.91), respectively, indicating the adequate discriminative ability of this prediction model. In the temporal validation, the c-statistics for the nomogram for predicting 3- and 5-year CSS were 0.89 (95% CI: 0.87–0.91) and 0.88 (95% CI: 0.83–0.94), respectively. The c-statistic was almost identical to the AUC of the ROC curve, which plotted the sensitivity (true positive rate) against 1-specificity (false positive rate) for the probability of an outcome (Figure 4). This finding implied the sufficient and stable discriminative ability of the prediction model. By comparing the nomogram-predicted and observed probabilities of 3- and 5-year CSS, the calibration was assessed with the calibration slope and Brier score, and visualized with calibration plots. The Brier score could also be used to reflect the overall performance of the model (22), with a score of 0 indicating perfect prediction accuracy and a score of 1 indicating poor prediction accuracy. In the internal validation, the optimism-adjusted Brier scores for the prediction of 3- and 5-year CSS were 0.09 (95% CI: 0.08–0.09) and 0.10 (95% CI: 0.10–0.11), respectively. In the temporal validation, the Brier scores were 0.08 (95% CI: 0.00–0.25) and 0.10 (95% CI: 0.00–0.38), respectively, and the calibration slope was 1.09, which is close to 1.0. The internal and temporal calibration curve demonstrated good consistency between the predicted and observed values of CSS (Figure 5).

Application of the nomogram
The nomogram could be used to predict the CSS of individual patients with UAC in clinical practice. For instance, a 50-year-old patient with a primary tumor of 45 mm in size (32 points), grade III (28 points), stage T2 (78 points), N0 (0 points), and M0 (0 points), who was treated with radical hysterectomy and lymph node dissection (0 points), had a total of 138 points. Correspondingly, the 3- and 5-year CSS probabilities were 71% and 63%, respectively. To allow for a more convenient estimate of CSS, the nomogram was incorporated into an online calculator with free access (https://betteryuan66.shinyapps.io/dynnomapp/), and a patient’s survival probability with 95% CIs could be derived by entering the values of the six variables and time. Utilizing the formula based on the model, the 5-year survival probability for the 15th patient in the development cohort was computed at 85%, which was close to the result (83%, 95% CI: 0.81–0.86) from the online calculator, which verified this calculator’s accuracy (Figure S2).
Additionally, the c-statistic of the nomogram was compared to those of the AJCC and SEER summary staging systems. The optimism-adjusted c-statistic of our nomogram for predicting CSS was 0.85 (95% CI: 0.84–0.86), which was higher than that of the AJCC (0.80, 95% CI: 0.79–0.81, P<0.001) and SEER summary staging systems (0.78, 95% CI: 0.77–0.79, P<0.001). These results indicated that the discriminative ability of this model was superior to that of the AJCC and SEER summary staging systems.
To further analyze the feasibility and validity of the prediction model, participants in the development cohort were stratified into high-risk and low-risk groups according to their prognostic scores derived from the model. The optimal cut-off value of risk-score was determined using the X-tile program (data not shown). Cases in the validation cohort were pooled with the same cut-off values of risk score, and the corresponding Kaplan-Meier survival curves were delineated respectively with significantly different prognosis (P<0.001), showing that patients at high risk of cancer-specific death could be identified through the model (Figure 6). The Kaplan-Meier survival curves showed that patients at high-risk exhibited a significantly poorer OS compared to those at low-risk.

Discussion
The identification of independent prognostic factors and the development of an accurate predictive model are crucial for optimal treatment planning, counselling, and follow-up for patients with UAC. By using SEER data, the present study identified tumor grade, stage T, stage N, stage M, tumor size, and surgery of the primary site to be independent prognostic factors of UAC, and constructed a novel nomogram for predicting the 3- and 5-year CSS of patients with the disease. The prediction model showed a satisfactory performance with accurate discrimination. After internal validation with bootstrap resampling and temporal validation, the model not only displayed a good discriminative ability, but also demonstrated an optimal agreement between the predicted and observed outcomes. Overall, the results of the present study showed that the model was both internally and temporally valid. Based on the model, each prognostic factor was quantified and visualized with the static nomogram so that patients’ 3- and 5-year UAC-specific survival probabilities could be predicted individually. For easier application of the model, we developed a web-based calculator (https://betteryuan66.shinyapps.io/dynnomapp/), from which a patient’s survival probability with 95% CI can be obtained after the values of six variables and time have been input. By using our prediction model, physicians will be able to judge individual risk based on a few known factors, predict the outcomes, and choose the therapies and follow-up for patients with UAC. Since patients classified as high risk have a higher likelihood of CSD, these patients should be administered comprehensive treatments and followed up closely.
To investigate the effect of treatment options on the prognosis of UAC, treatment modalities such as surgery, chemotherapy, and radiotherapy were enrolled as candidate variables. The results verified surgery of the primary tumor to be an independent beneficial factor. Incorporating therapeutic factors and tumor size and grade, the prognostic nomogram had added predictive value compared to the AJCC and SEER summary staging systems, which indicated that it could play a supplementary role. Combined treatments were not analyzed due to the number of cases in different groups being obviously different. In the development cohort, 2,894 (51.1%) patients received primary surgery, 671 (11.8%) received primary surgery and radiotherapy, 107 (1.8%) underwent primary surgery and chemotherapy, 940 (16.6%) underwent primary surgery and CCRT, 595 (10.5%) underwent CCRT, 297 (5.2%) received radiotherapy only, 35 (0.6%) received chemotherapy only, and 116 (2.0%) did not receive any treatment. Consequently, the present study has provided a basis for further research using a greater sample size.
To our knowledge, this was the first study to develop a prognostic model for UAC. A few nomograms have been developed for cervical cancer overall; however, due to its rarity, UAC has not been a specific focus of studies to date. Polterauer et al.’s study (23) included 528 participants with invasive cervical cancer after surgical staging and developed a nomogram to predict 3- and 5-year OS. Their study involved 86 (16.3%) patients with adenocarcinoma, and the estimated optimism-corrected c-statistic of the model was 0.723 for the prediction of OS. Zhou et al. (12) retrospectively analyzed 1,563 patients with surgically treated stage IA–IIB cervical cancer and established a nomogram to predict 5-year OS with a c-statistic of 0.71. In this population, a total of 168 (10.7%) participants were with non-squamous carcinoma. In Lee’s study involving 10 centers (24), 1,702 patients with stage IB–IIA cervical cancer who were treated with radical hysterectomy followed by adjuvant radiotherapy were analyzed, and a nomogram to predict 5-year OS was developed with a c-statistic of 0.69. Based on an analysis of 209 patients with stage IB2–IVA cervical cancer treated with definitive CCRT, Shim et al. (25) constructed a nomogram to predict the 5-year OS, and the c-statistic was 0.69. Utilizing SEER data from between 1988 and 2005, Galic et al. (26) explored the effect of tumor histology on the outcomes of women with cervical carcinoma and concluded that both early- and advanced-stage UAC had inferior survival to SCC. As revealed in these studies, histologic type was a vital independent prognostic factor for 5-year OS prediction.
A fundamental distinction of UAC from squamous lesions is its more aggressive behavior and worse prognosis. The present study, which included 5,655 UAC patients from the SEER database, constructed a prediction nomogram for 3- and 5-year CSS with optimism-adjusted c-statistics of 0.90 (95% CI: 0.89–0.91) and 0.89 (95% CI: 0.89–0.91), respectively. This prediction nomogram was internally and temporally validated. The c-statistic, which ranges from 0.5 to 1.0, is commonly used to assess the discriminative ability of a model, and a larger c-statistic suggests higher accuracy for distinguishing patient outcomes. Therefore, the present model displayed good accuracy and temporal validity for predicting CSS in UAC patients.
As well as histologic type, several factors, including age, FIGO stage, tumor size, stromal invasion, lympho-vascular space invasion (LVSI), parametrial involvement, and the use of concurrent chemotherapy, have been identified in previous studies on cervical carcinoma and incorporated into prediction models (12,23-25). In our study with UAC, the following were identified as independent prognostic factors and subsequently incorporated into the model: tumor grade, stage T, stage N, stage M, size of the tumor, and surgery of the primary site, which was consistent with the findings of other studies (7-9,27). In the nomogram, stage T made the largest contribution to prognosis, followed by tumor size and tumor grade. According to the NCCN guidelines, tumor grade was not listed as one of the high- or intermediate risk factors for SCC (4). In our study, tumor grade II (moderately differentiated), grade III and IV (poorly differentiated or undifferentiated) increased the risk of poor prognosis (HR =1.41, 95% CI: 1.20–1.67, P<0.0001; HR =2.10, 95% CI: 1.78–2.48, P<0.0001, respectively). Baalbergen et al. (7) retrospectively analyzed 305 cases with UAC and found FIGO stage, tumor grade, and lymph node metastases to be significant prognostic factors for survival. Gadducci et al. (28) concluded that the most investigated clinical-pathological prognostic factors for UAC were FIGO stage, lymph nodal status, tumor size, tumor grade, patient age, depth of cervical invasion, LVSI, and parametrial involvement. Tumor size was established as another independent prognostic factor, although it is also a determining factor of AJCC stage T1a-2a. Our study incorporated treatment strategies into the predictive variables for the Cox hazard regression analysis, which was distinct from other studies. As shown in Table S2, cases who received surgery of the primary site showed improved outcomes (HR =0.56, 95% CI: 0.49–0.63, P<0.001). The results suggested that increased tumor burden (tumor size and TNM stage) and tumor biological behavior (histologic grade) indicated poor prognosis, whereas surgery of the primary site was associated with improved outcomes. Regarding the association between the number of lymph nodes involved and the prognosis, no significant difference was found in the multivariate Cox regression analysis of our study. Zhou et al. (29) retrieved data of 312 UAC patients with stage I–IIB and found that the number of positive pelvic nodes and age at surgery were independent prognostic factors for OS, while postoperative tumor diameter (≥4 cm) and the number of positive nodes were independent prognostic factors for relapse-free survival (RFS). The discrepancy in results may be attributable to the following reasons. First, 816 cases for whom the number of positive lymph nodes was missing were at an advanced stage, including 327 (40%) with AJCC stage III and 359 (43.9%) with AJCC stage IV. Among these cases, 742 (90.9%) presented with LN involvement and 74 (9.1%) without LN involvement. If the cases with missing data were deleted, the conclusions would have been biased and not applicable to the UAC population as a whole. To control bias and to reach valid conclusions for the target cohort, we analyzed the relationship between missing data on the number of lymph nodes examined and those on the number of lymph nodes involved, and assumed the data were missing at random. The missing data were handled with a multiple imputation method by chained equations (19). In the procedure, the risk of bias due to missing data was minimized rather than fully eliminated, because full elimination could have produced different results. Second, >20 histologic subtypes of UAC were included in the analyses, and the heterogeneity of histology could also have influenced the outcomes. Therefore, prospective randomized studies are further warranted.
By using the SEER database, the present study developed a novel predictive model for UAC patients, which was verified as a more individualized tool than the AJCC staging system for evaluating the prognosis of UAC. However, the present study had several limitations. First, the model was based on retrospective analysis, which may have caused biases due to the lack of random assignment, patient selection, and some missing values. Second, information on some potential independent prognostic variables, such as parametrial involvement, margin status, stromal invasion, and LVSI, were unavailable from the SEER database, which could also have increased the performance index of the model. Since the information of the database spans nearly 30 years, the details of adjuvant treatment have varied, which may appear as a confounding factor for predicting prognosis (30,31). Third, although the prediction model has been internally and temporally validated with the SEER database, more institutions, especially those outside of the United States, are encouraged to validate the model externally before it is applied to the general population.
Conclusions
In conclusion, the present study identified tumor grade, stage T, stage N, stage M, tumor size, and surgery of the primary site as independent prognostic factors and constructed a novel nomogram to predict 3- and 5-year CSS for UAC patients. The nomogram displayed a good discriminative ability and an optimal accuracy. In the form of a static nomogram or an online prediction tool (available at https://betteryuan66.shinyapps.io/dynnomapp/), this nomogram could assist clinicians to quantify the risk of mortality, optimize the treatment plan, and personalize follow-up for UAC patients in an effective and convenient way. Large prospective multicenter studies are warranted for additional external validation, generalization, and application of this prediction model.
Acknowledgments
The authors would like to thank Dr. Jinhui Liu from the First Affiliated Hospital of Nanjing Medical University for the direction of statistical analysis and Ms. Zhiwei Zhang from Nanjing Medical University for polishing the manuscript.
Funding: The study was supported by the Project of Maternal and Child Health of Jiangsu, China (F201844) (CL). The funders had no role in the design of study, data collection, analysis, interpretation, or in the preparation of the manuscript.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-6201
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-6201
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-6201). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Islami F, Fedewa SA, Jemal A. Trends in cervical cancer incidence rates by age, race/ethnicity, histological subtype, and stage at diagnosis in the United States. Prev Med 2019;123:316-23. [Crossref] [PubMed]
- Abu-Rustum NR, Yashar, Catheryn M, et al. National Comprehensive Cancer Network (NCCN) Clinical Practice Guidelines in Oncology Cervical Cancer, Version 2.0, 2020. Available online: https://www.nccn.org/professionals/physician_gls/pdf/cervical.pdf.
- Bhatla N, Berek JS, Cuello Fredes M, et al. Revised FIGO staging for carcinoma of the cervix uteri. Int J Gynaecol Obstet 2019;145:129-35. [Crossref] [PubMed]
- Hu K, Wang W, Liu X, et al. Comparison of treatment outcomes between squamous cell carcinoma and adenocarcinoma of cervix after definitive radiotherapy or concurrent chemoradiotherapy. Radiat Oncol 2018;13:249. [Crossref] [PubMed]
- Baalbergen A, Ewing-Graham PC, Hop WC, et al. Prognostic factors in adenocarcinoma of the uterine cervix. Gynecol Oncol 2004;92:262-7. [Crossref] [PubMed]
- Park JY, Kim DY, Kim JH, et al. Outcomes after radical hysterectomy in patients with early-stage adenocarcinoma of uterine cervix. Br J Cancer 2010;102:1692-8. [Crossref] [PubMed]
- Khalil J, Bellefqih S, Afif M, et al. Prognostic factors affecting cervical adenocarcinoma: 10 years experience in a single institution. Arch Gynecol Obstet 2015;292:915-21. [Crossref] [PubMed]
- Yin J, Yang Y, Ma K, et al. Clinicopathological characteristics and prognosis of pulmonary pleomorphic carcinoma: a population-based retrospective study using SEER data. J Thorac Dis 2018;10:4262-73. [Crossref] [PubMed]
- He J, Albertsen PC, Moore D, et al. Validation of a Contemporary Five-tiered Gleason Grade Grouping Using Population-based Data. Eur Urol 2017;71:760-3. [Crossref] [PubMed]
- Zhou H, Li X, Zhang Y, et al. Establishing a Nomogram for Stage IA-IIB Cervical Cancer Patients after Complete Resection. Asian Pac J Cancer Prev 2015;16:3773. [Crossref] [PubMed]
- Han DS, Suh YS, Kong SH, et al. Nomogram predicting long-term survival after d2 gastrectomy for gastric cancer. J Clin Oncol 2012;30:3834-40. [Crossref] [PubMed]
- Surveillance Research Program. National Cancer Institute SEER*Stat software, 8.3.6, 2020. National Cancer Institute, National Institutes of Health. Available online: http://seer.cancer.gov/seerstat.
- SEER*Stat Database: Incidence - SEER Research Data, 9 Registries, Nov 2019 Sub (1975-2016)- Linked To County Attributes- Time Dependent (1990-2016) Income/Rurality, 1969-2016 Counties, National Cancer Institute, DCCPS, Surveillance Research Program, released April 2019, based on the November 2018 submission. Available online: www.seer.cancer.gov.
- Carcangiu M, Kurman RJ, Carcangiu ML, et al. WHO classification of tumours of female reproductive organs. 4th ed. Lyon: International Agency for Research on Cancer; 2014. p.171.
- Jack A, Percy C, Sobin L, et al. International classification of diseases for oncology: ICD-O. 3rd ed. World Health Organization; 2000. p.56-62.
- World Medical Association. World Medical Association declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA 2013;310:2191-4. [Crossref] [PubMed]
- Mühlenbruch K, Kuxhaus O, di Giuseppe R, et al. Multiple imputation was a valid approach to estimate absolute risk from a prediction model based on case-cohort data. J Clin Epidemiol 2017;84:130-41. [Crossref] [PubMed]
- Kwon J, Eom KY, Kim YS, et al. The Prognostic Impact of the Number of Metastatic Lymph Nodes and a New Prognostic Scoring System for Recurrence in Early-Stage Cervical Cancer with High Risk Factors: A Multicenter Cohort Study (KROG 15-04). Cancer Res Treat 2018;50:964-74. [Crossref] [PubMed]
- Camp RL, Dolled-Filhart M, Rimm DL. X‐tile: a new bio‐informatics tool for biomarker assessment and outcome‐based cut‐point optimization. Clin Cancer Res 2004;10:7252-9. [Crossref] [PubMed]
- Steyerberg EW, Vickers AJ, Cook NR, et al. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology 2010;21:128-38. [Crossref] [PubMed]
- Polterauer S, Grimm C, Hofstetter G, et al. Nomogram prediction for overall survival of patients diagnosed with cervical cancer. Br J Cancer 2012;107:918-24. [Crossref] [PubMed]
- Lee HJ, Han S, Kim YS, et al. Individualized prediction of overall survival after postoperative radiation therapy in patients with early-stage cervical cancer: a Korean Radiation Oncology Group study (KROG 13-03). Int J Radiat Oncol Biol Phys 2013;87:659-64. [Crossref] [PubMed]
- Shim SH, Lee SW, Park JY, et al. Risk assessment model for overall survival in patients with locally advanced cervical cancer treated with definitive concurrent chemoradiotherapy. Gynecol Oncol 2013;128:54-9. [Crossref] [PubMed]
- Galic V, Herzog TJ, Lewin SN, et al. Prognostic significance of adenocarcinoma histology in women with cervical cancer. Gynecol Oncol 2012;125:287-91. [Crossref] [PubMed]
- Chen J, Wang R, Zhang B, et al. Safety of ovarian preservation in women with stage I and II cervical adenocarcinoma: a retrospective study and meta-analysis. Am J Obstet Gynecol 2016;215:460.e1-e13. [Crossref] [PubMed]
- Gadducci A, Guerrieri ME, Cosio S. Adenocarcinoma of the uterine cervix: Pathologic features, treatment options, clinical outcome and prognostic variables. Crit Rev Oncol Hematol 2019;135:103-14. [Crossref] [PubMed]
- Zhou J, Chen Y, Xu X, et al. Postoperative clinicopathological factors affecting cervical adenocarcinoma: Stages I-IIB. Medicine (Baltimore) 2018;97:e9323. [Crossref] [PubMed]
- Noone AM, Lund JL, Mariotto A, et al. Comparison of SEER treatment data with Medicare claims. Med Care 2016;54:e55-64. [Crossref] [PubMed]
- Yu JB, Gross CP, Wilson LD, et al. NCI SEER public-use data: applications and limitations in oncology research. Oncology (Williston Park) 2009;23:288-95. [PubMed]



