
Germline and tumor BRCA1/2 mutations in Chinese high grade serous ovarian cancer patients
Introduction
Ovarian cancer is the third malignancy of the female genital tract, following cervical and uterine cancer. In 2020, there were 21,750 estimated new diagnoses of ovarian cancer and 13,940 deaths from the disease in the United States; deaths were higher than from cancer of the uterine corpus and cervix (1). High grade serous ovarian cancer (HGSOC) is the most common histological subtype of ovarian cancer, which is characterized by a high risk of recurrence and frequent genetic and epigenetic alterations of homologous recombination (HR) pathway genes, accounting for 70-80% of deaths from ovarian cancers (2-4). HR deficiency can be assessed by the presence of germline and somatic mutations in HR genes, including the breast cancer susceptibility genes 1 and 2 (BRCA1 and BRCA2), Fanconi anemia genes (BRIP1, PALB2), the core RAD genes (RAD51C, RAD51D), and genes involved in HR pathways either directly (CHEK2, BARD1, NBN, ATM) or indirectly (CDK12) (5). The lifetime risk of epithelial ovarian cancer of well-established moderate- and high-penetrance susceptibility genes varies approximately between 5% and 60%, among which BRCA1/2 genes had the highest risk coefficient (5). Mutations in BRCA1/2 genes may lead to chromosomal instability, promote cell proliferation and prevent differentiation of normal cells, thus resulting in the occurrence and development of tumor (6). It is also an important biomarker for therapeutic intervention, as ovarian cancer patients with germline or somatic BRCA1/2 mutation benefit more from chemotherapy and poly (ADP-Ribose) polymerase (PARP) inhibitors than non-carriers (7-10). Therefore, fully understanding BRCA1/2 mutations is of great significance for clinical genetic counseling and guiding medical management strategies of ovarian cancer.
Common mutation types include single-nucleotide variants (SNV), insertion/deletion (indel) and large genomic rearrangement (LRG) and can occur in both germ cells and somatic cells. To our knowledge, due to the insufficient precision of bioinformatics algorithm on copy number variation in next-generation sequencing (NGS) method, previous studies on the prevalence of BRCA1/2 mutation in ovarian cancer mainly evaluated germline SNV and indel, but usually lacked LGR detection (11-17). Besides that, studies on tumor BRCA1/2 mutation are still limited for the relatively lower prevalence and more complex detection workflow (4,18,19). Although some studies investigated BRCA1/2 mutations in ovarian cancer patients in our country, none of them has simultaneously clarified the overview of germline and tumor BRCA1/2 mutation including SNV, indel and LGR, also none specifically in HGSOC patients. In this study, we took advantage of NGS and multiplex ligation-dependent probe amplification (MLPA) technologies to investigate deleterious germline and somatic BRCA1/2 mutations in Chinese HGSOC patients, and analyze their association with clinicopathological factors and survival outcomes. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-6827).
Methods
Study patients and samples
An unselected cohort of 115 Chinese HGSOC patients undergone surgery in Fudan University Shanghai Cancer Center from 2014 to 2015 were sequentially selected for our study population. Patients enrolled were supposed to meet the following inclusion criteria: (I) pathologically confirmed HGSOC at the Department of Pathology of Fudan University Shanghai Cancer Center; (II) sufficient formalin-fixed paraffin-embedded (FFPE) sections for somatic testing; (III) DNA extracted was qualified for NGS and MLPA testing; and (IV) willingness to provide signed consent in advance of the trial. Patients not meeting all of these inclusion criteria were excluded. Blood and paired FFPE tissue samples were collected and retrieved from our biobank and the Department of Pathology for BRCA1/2 testing. Genomic DNA extracted from tumor tissue sections and peripheral blood were performed using QIAamp DNA MiniKit and QIAamp DNA MidiKit (QIAgen, Valencia, CA), respectively. Clinicopathological parameters were electronically retrieved from the Hospital Information System (HIS) of Fudan University Shanghai Cancer Center. Median age at diagnosis in this cohort was 51 years (range, 38–79 years). All patients were followed up until July, 2020, or death. The median follow-up time was 44.8 (0.2–73.2) months. Disease relapse or progression was determined by medical imaging, serology, or histology. Progression-free survival (PFS) was measured from diagnosis to local or systemic recurrence or the last follow-up, while OS was measured from diagnosis to death or the last follow-up. Patients with one or more family members within two generations with breast or ovarian cancer were considered as having an HBOC family history. No kinship was found among them according to information of genetic counseling. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Medical Ethics Committee of Fudan University Shanghai Cancer Center (NO.: FWA00030121) and informed consent was taken from all individual participants.
BRCA1/2 SNV/indel detection by NGS
For targeted NGS analysis, total 265 primer pairs in two pools (133 pairs in pool 1, and 132 pairs in pool 2) were used in Oncomine™ BRCA Research Assay (Thermo Fisher Scientific, Schaumburg, IL, USA), which can amplify the entire coding regions and 20 bp upstream or downstream of exon–intron boundaries of BRCA1/2 genes. Multiplex PCR was performed using 20 ng genomic DNA with the following cycling conditions: 99 °C × 2 minutes, 20 cycles of 99 °C × 15 seconds, and 60 °C × 4 minutes. The amplicons were treated with 2 µL FuPa reagent to partially digest primers and phosphorylate the amplicons with the following conditions: 50 °C × 10 minutes, 55 °C × 10 minutes, and 60 °C × 20 minutes. The diluted barcodes (Thermo Fisher Scientific) were ligated with the following conditions: 22 °C × 30 minutes, and 68 °C × 5 minutes, and 72 °C × 10 minutes. Libraries were purified using Agencourt AMPure XP reagents (Beckman Coulter, Brea, CA, USA). Concentration was measured using an Ion Library Quantitation Kit (Thermo Fisher Scientific), then the same amount of 100 pmol/L libraries was pooled in one sequencing reaction. Emulsion PCR was implemented with the Ion OneTouch™ 2.0 System and Hi-Q™ View OT2 reagents (Thermo Fisher Scientific) following the manufacturer’s instructions. The template-positive particles were purified using Ion OneTouch™ ES system and MyOne™ Streptavidin C1 Beads (Thermo Fisher Scientific). Parallel sequencing was performed on a Personal Genome Machine (PGM) sequencer using the Ion PGM™ Hi-Q™ Sequencing Kit according to the manufacturer’s instructions. Sequencing was performed using 500 flow runs that generated ~200 bp reads.
The sequence data were processed using standard pipeline on Torrent Suite™ version 5.4 (Thermo Fisher Scientific) as previously described (20). Annotations including SNV, indel, and splice site alteration were performed using Ion Reporter™ version 5.4 (Thermo Fisher Scientific). Binary alignment map (BAM) files were visually confirmed with the Integrative Genomics Viewer (IGV) 2.4.4. Error artifacts of sequence, alignment, or variant call were discarded. Minor allele frequency (MAF) of variant less than 0.01 was considered for further pathogenicity evaluation. Variants were classified as pathogenic, likely pathogenic, uncertain significance, likely benign and benign according to ACMG guideline (21). Pathogenic/likely pathogenic (P/LP) variants were regarded as deleterious mutations with clinical significance. MAF was identified from population database including 1000 Genomes Project database (http://phase3browser.1000genomes.org/) and dbSNP (https://www.ncbi.nlm.nih.gov/snp/) and Exome Aggregation Consortium and Exome Sequencing Project. The variants pathogenic determination referred to databases such as the BRCA Exchange database (https://brcaexchange.org/favicon.ico), LOVD database (https://databases.lovd.nl/shared/genes) and ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/) and published papers. Bioinformatic tools including SIFT (http://sift.jcvi.org), Align GVGD (http://agvgd.iarc.fr/agvgd_input.php) and PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2) were used as supplementary evidence to prove that a variant may affect normal function.
LGRs detection by MLPA
Genomic DNA extracted from the patient’s peripheral blood was used for BRCA LGRs detection following the MLPA instructions (MRC-Holland, Amsterdam, The Netherlands). Probe mix P002 and P087 were used for the detection or confirmation of BRCA1 LGRs. Similarly, probe mix P045 and P090 were used for BRCA2 LGRs. Briefly, 100 ng genomic DNA dissolved in 5 µL TE buffer was used for denaturation with 98 °C × 5 minutes, then MLPA probe mix was added for hybridization with the following conditions: 95 °C × 1 minute, and 60 °C × 16–20 hours. After hybridization, a Ligase-65 master mix was added for ligation with the following conditions: 54 °C pause, 54 °C × 15 minutes, and 98 °C × 5 minutes. Multiplex PCR was performed with the following cycling conditions: 35 cycles of 95 °C × 30 seconds, 60 °C × 30 seconds, 72 °C × 60 seconds and an additional 72 °C × 20 minutes. Finally, electrophoresis and data analysis were performed on ABI 3500 machine (Thermo Fisher Scientific) and Coffalyser.NET software (MRC-Holland).
Statistical analysis
Pathologic characteristics tabulated by their types or ranges were compared between groups by chi-square test or Fisher’s exact test, as appropriate. Survival analysis was performed using log-rank test, Kaplan-Meier analysis and multivariate Cox proportional hazards regression models. All analyses were performed using GraphPad Prism 8.0 (GraphPad Software Inc., San Diego, USA) and SPSS version 19 (SPSS version 19; SPSS, Chicago, IL). P values <0.05 were considered as statistically significant.
Results
Germline and somatic BRCA1/2 mutations in HGSOC
The complete list of deleterious mutations identified in this cohort and their frequency were summarized in Table 1 and Figure 1. There were 25 (21.7%) germline BRCA1/2 mutations found by NGS testing, including 19 (16.5%) BRCA1 and 6 (5.2%) BRCA2 mutations (Figure 1A). In addition, 5 (4.3%) BRCA1 LGRs were detected in blood DNA by MPLA, whereas no BRCA2 LGRs were found in these patients (Figure 1A). The entire germline BRCA1/2 mutation frequency was 26.1%. Among them, the proportion of frameshift, LGRs, nonsense, splice site alteration and nonsense were 50.0%, 16.7%, 16.7%, 10.0% and 6.6%, respectively (Figure 1B). Moreover, c.4228delG, c.1786delC, c.3059delC, c.2341G>T found in BRCA1 and c.6382_6386delAAAGA, c.5851dupA found in BRCA2 were novel that hasn’t been reported in database (Table 1).
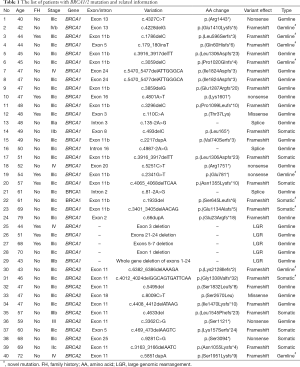
Full table
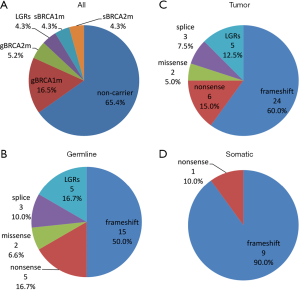
All germline mutations detected by NGS were also found in paired tumor tissues. In addition, 10 (8.6%) mutations were found only in tumor tissues, including 5 (4.3%) in BRCA1 and 5 (4.3%) in BRCA2 (Figure 1A). Nine (90%) of them were frameshift and the other one was nonsense (Figure 1D). c.1933del, c.3401_3405delAACAG found in BRCA1 and c.4012_4024delGGCAGTGATTCAA found in BRCA2 were not reported before. The mutational allele frequency ranged from 7.4% to 45.5%. Since LGRs detected in blood DNA theoretically exists in tumor tissues, the entire tumor BRCA1/2 mutation frequency was 34.8%. Among these tumor BRCA1/2 mutations, the proportion of frameshift, LGRs, nonsense, splice site alteration and nonsense were 60.0%, 12.5%, 15.0%, 7.5% and 5.0%, respectively (Figure 1C).
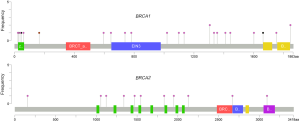
All BRCA1/2 mutations including SNV, indel and LGRs were visualized by IGV software or Coffalyser software. Notably, no patient in this cohort was found with two or more deleterious mutations showing a mutual exclusion of BRCA1/2 mutation. In terms of distribution, all mutations are scattered in various functional domains and protein binding regions of BRCA1/2 (Figure 2).
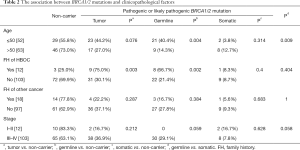
The association between BRCA mutations and clinicopathological factors
Age at diagnosis, FIGO stage, family history of HBOC and family history of other cancer than breast and ovarian cancer, including esophageal cancer, gastric cancer, liver cancer, lung cancer, pancreatic cancer, intestinal cancer and malignant teratoma, were included in pathologic characteristics. The median age in this cohort was 51 years (range, 38–79 years). 10.4% of these patients had potentially significant family history of HBOC. The proportion of germline (66.7%) and tumor (75%) mutation carriers was significantly increased for patients with family history when compared with those without (P<0.05), while somatic BRCA1/2 mutation carriers showed no significant association with inheritance patterns (8.3%, P=0.4, Table 2). In addition, patients with deleterious germline BRCA1/2 mutation appeared to be younger than non-carriers (mean age, 50.9 years vs. 54.4 years, P=0.004), while somatic or tumor BRCA1/2 mutation carriers showed no significant association with ages (mean age, 58.7 vs. 54.4 years for somatic, P=0.314, 52.9 years vs. 54.4 years for tumor, P=0.076, Table 2). Notably, the onset age of patients with germline mutations was significantly younger than that of patients with somatic mutations (mean age, 50.9 vs. 58.7 years, P=0.009). No significant association was found between BRCA1/2 status and clinicopathological factors including stage and family history of other cancers. The relationships between BRCA1 and BRCA2 mutation and clinicopathological factors are listed in Table 3. No statistical significance was found in both ages (mean age, 50.5 vs. 52.5 years, respectively, P=1, Table 3) and family history of HBOC (P=0.155) between the two groups.
Full table
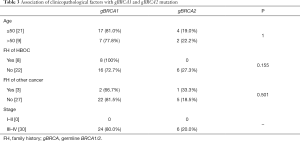
Full table
Survival analysis
At a median follow-up of 44.8 months (range, 0.2–73.2 months), there were 62 (53.9%) recurrences and 52 (45.2%) deaths in this cohort. The Kaplan Meier and Cox regression analysis for PFS and OS by mutation status are shown in Figure 3 and Figures S1,S2. In univariate analysis, patients with tumor BRCA1/2 mutations had significant improvements than non-carriers in OS in the first two years after surgery, with HR 22.322 (95% CI, 21.52–23.123, P=0.018). The results were confirmed by multivariate analysis with HR 0.35 (95% CI, 0.07–1.64, P=0.022) for germline mutations and HR 0.2 (95% CI, 0.04–0.9, P=0.008) for tumor mutations (Figure S2). No significant impacts were found between various mutation status in PFS outcomes. No significant differences were found between BRCA1 or BRCA2 and non-carriers in PFS or OS (Figure 4). The low number of events precluded survival analysis of somatic subgroups.
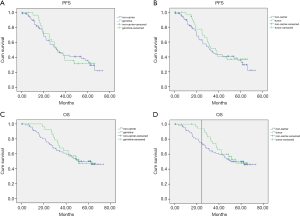
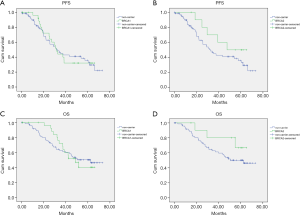
Discussion
This study specifically focused on Chinese HGSOC patients, and firstly simultaneously clarified the overview of germline and somatic BRCA1/2 mutations including SNV, indel and LGR, and further investigated the relationship between BRCA1/2 mutation with clinical pathologic characters and survival outcomes.
Current findings showed that the overall incidence of deleterious BRCA1/2 mutations in tumor of HGSOC patients was 34.8%, of which germline mutation accounted for the vast majority, and the proportion was 26%. However, this frequency varies greatly in the reports of other countries. One study on germline BRCA1/2 mutation in 1915 unselected ovarian carcinomas was conducted in the United States, they found the deleterious mutation frequency including SNV, indel and LGRs in 1498 HGSOC patients was 16.0% by targeted capture (11), while another study from the United States showed that 98 out of 433 (22.6%) HGSOC patients were identified with germline BRCA1/2 mutation (SNV, indel and LGRs) by sequencing and MLPA (12). In a prospectively study mainly focused on the Arab population, a prevalence of 25.7% deleterious germline BRCA1/2 mutation (SNV and indel) was found in 74 HGSOC patients by sequencing (13). Among Asian countries, a relatively small number screening was performed for germline BRCA1/2 mutation (SNV and indel) in 230 unselected ovary cancer patients by targeted panel sequencing, they found 22 out of 74 (29.7%) HGSOC patients carrying deleterious mutation (SNV, indel and LGRs) and the result was consistent with a respective study in Japan that reported 28.5% of germline BRCA1/2 mutation (SNV and indel) by NGS testing (14,15). On the other hand, one study from Thailand found the germline BRCA1/2 mutation (SNV and indel) frequency in HGSOC was 25.7% by NGS testing (16). In China, a nationwide multicenter germline BRCA1/2 mutation prevalence study was conducted previously, 601 HGSOC patients were enrolled in this study, they found the deleterious BRCA1/2 mutation prevalence (SNV, indel and LGRs) was 30.9% by targeted DNA sequencing, which was the high boundary of the previously reported range (17). Although the similar sequencing method for analysis of germline BRCA1/2 mutations including SNV, indel and LGRs was used in these studies, the incidence possibly varied in differences with geography, ethnicity, population or even economic level. In general, the BRCA1/2 mutation frequencies in Asian countries seems to be higher than that in Western countries. The deleterious germline BRCA1/2 mutant rate in our study was similar to some of the above work (13,16). In addition, we identified 4.3% germline BRCA1 mutation as LGRs, which suggests LGRs detection should be considered in mutation screening and genetic counseling in HGSOC patients, especially for young patients without SNV and indel.
Beyond germline mutations, somatic mutations in HR pathway have been implicated in sporadic ovarian cancer, accounting for approximately 6% of HGSOC (22). Our results showed that the somatic BRCA1 and BRCA2 gene mutation frequencies in Chinese HGSOC patients were 4.3% and 4.3% respectively, which was slightly higher than the previously reported frequencies of 3% and 3% respectively in Western population (4). Currently in China, early screening and routine examination of BRCA1/2 mutation for ovarian cancer patients is still incomplete, a higher percentage of patients diagnosed with distant metastasis, which may lead to the deviation of somatic mutation frequency. In this study, tumor samples showed high proportion of III and IV FIGO stage as shown before, these might be the reasons of higher somatic BRCA1/2 mutation frequencies we observed.
From the clinical point of view, the PARP inhibitors efficacy includes both germline mutant and sporadic ovarian cancers with HR deficiency (23). Moreover, an additional important issue of current research is to expand their use in ovarian cancers with HR deficiency or even beyond HR-deficient. Recently, novel combinations of PARP inhibitors with drugs that inhibit HR, such as anti-angiogenics, immune checkpoint, PI3K/AKT/mTOR, WEE1, MEK, and CDK4/6 inhibitors that have also been proposed in patients with de novo or acquired HR proficiency to PARP inhibitors (24). However, further clinical practices are required to reduce overlapping toxicities by optimizing dose and schedule, and to utilize the combinations to highly selected patients who would not otherwise benefit from single PARP inhibitors (24). Meanwhile, a comprehensive mutation detection capability would accelerate the identification of cancers appropriate for combined therapy.
Patients with germline BRCA1/2 mutation are associated with HBOC syndrome, which is characterized by familial clustering of breast and ovarian cancers (25). In this study, we observed significant associations between BRCA1/2 mutation status and HBOC family history. In patients with family history, the deleterious germline BRCA1/2 mutation incidence was as high as 66.7%. This result is in concordance with studies that showed 60–70% of patients with family history who had BRCA1/2 mutation (17,26). Nonetheless, we also observed the deleterious germline BRCA1/2 mutation rate was 21.4% in patients without HBOC family history in our study. In addition to family history, we also observed approximately 40% of HGSOC patients under 50 years had deleterious germline BRCA1/2 mutation, which suggests onset age under 50 years is another clinical factor associated with germline BRCA1/2 status. This finding is consistent with the previous research on young Israeli women which reported about 50% (27). These data conclude the probability to find a mutant rate varies greatly in different clinical subgroups, which leads to the hypothesis that testing for BRCA1/2 mutations addressed within each specific clinical scenario could be more cost-effective for patients. A recently study also suggested that there is no evidence that delivering a widespread BRCA1/2 testing for ovarian cancer patients is cost-effective with respect to standard practice for preventive and therapeutic purposes (28). Therefore, how to select candidates for BRCA1/2 testing is a feasible and challenging work in the future.
In our study, we did not find significant impacts of germline and tumor BRCA1/2 mutations on PFS, but tumor and germline BRCA1/2 mutations were all inclined to better outcomes on OS. However, the advantage on OS for those with BRCA1/2 mutations compared to non-carriers showed a time-dependent decline, as in previous report (11). Specifically, we found this advantage was more significant within 2 years after surgery. The improved survival outcomes were likely caused by a higher sensitivity to platinum in patients with BRCA1/2 mutations than non-carriers (29).
In summary, there is a high germline (26.0%) and tumor (34.8%) BRCA1/2 mutation incidences including SNV, indel and LGR in Chinese HGSOC patients. Germline mutations were associated with HBOC family history and age at diagnosis of HGSOC, whereas somatic mutations were not. In our study, tumor BRCA1/2 mutations predicted increased sensitivity to platinum-based chemotherapy and significantly improved survival outcomes, but the advantage was time-depended. A larger cohort and even multigene panel testing should be examined to clarify the relation between BRCA1/2 mutation and prognosis.
Acknowledgments
Funding: This study was supported by Innovation Group Project of Shanghai Municipal Health Commission (Project No: 2019CXJQ03), Shanghai Science and technology development fund (Project No: 19MC1911000) and Shanghai Municipal Key Clinical Specialty (shslczdzk01301).
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/atm-20-6827
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-6827
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-6827). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Medical Ethics Committee of Fudan University Shanghai Cancer Center (NO.: FWA00030121) and informed consent was taken from all individual participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Bowtell DD, Bohm S, Ahmed AA, et al. Rethinking ovarian cancer II: reducing mortality from high-grade serous ovarian cancer. Nat Rev Cancer 2015;15:668-79. [Crossref] [PubMed]
- Torre LA, Trabert B, DeSantis CE, et al. Ovarian cancer statistics, 2018. CA Cancer J Clin 2018;68:284-96. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609-15. Erratum in: Nature 2012;490:298. [Crossref] [PubMed]
- Boussios S, Mikropoulos C, Samartzis E, et al. Wise Management of Ovarian Cancer: On the Cutting Edge. J Pers Med 2020;10:41. [Crossref] [PubMed]
- Tutt A, Ashworth A. The relationship between the roles of BRCA genes in DNA repair and cancer predisposition. Trends Mol Med 2002;8:571-6. [Crossref] [PubMed]
- Papa A, Caruso D, Strudel M, et al. Update on Poly-ADP-ribose polymerase inhibition for ovarian cancer treatment. J Transl Med 2016;14:267. [Crossref] [PubMed]
- Candido-dos-Reis FJ, Song H, Goode EL, et al. Germline mutation in BRCA1 or BRCA2 and ten-year survival for women diagnosed with epithelial ovarian cancer. Clin Cancer Res 2015;21:652-7. [Crossref] [PubMed]
- Coleman RL, Oza AM, Lorusso D, et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017;390:1949-61. [Crossref] [PubMed]
- Ledermann J, Harter P, Gourley C, et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol 2014;15:852-61. [Crossref] [PubMed]
- Norquist BM, Harrell MI, Brady MF, et al. Inherited Mutations in Women With Ovarian Carcinoma. JAMA Oncol 2016;2:482-90. [Crossref] [PubMed]
- Alsop K, Fereday S, Meldrum C, et al. BRCA mutation frequency and patterns of treatment response in BRCA mutation-positive women with ovarian cancer: a report from the Australian Ovarian Cancer Study Group. J Clin Oncol 2012;30:2654-63. [Crossref] [PubMed]
- Ashour M, Ezzat Shafik H. Frequency of germline mutations in BRCA1 and BRCA2 in ovarian cancer patients and their effect on treatment outcome. Cancer Manag Res 2019;11:6275-84. [Crossref] [PubMed]
- Hirasawa A, Imoto I, Naruto T, et al. Prevalence of pathogenic germline variants detected by multigene sequencing in unselected Japanese patients with ovarian cancer. Oncotarget 2017;8:112258-67. [Crossref] [PubMed]
- Enomoto T, Aoki D, Hattori K, et al. The first Japanese nationwide multicenter study of BRCA mutation testing in ovarian cancer: CHARacterizing the cross-sectionaL approach to Ovarian cancer geneTic TEsting of BRCA (CHARLOTTE). Int J Gynecol Cancer 2019;29:1043-9. [Crossref] [PubMed]
- Manchana T, Phoolcharoen N, Tantbirojn P. BRCA mutation in high grade epithelial ovarian cancers. Gynecol Oncol Rep 2019;29:102-5. [Crossref] [PubMed]
- Wu X, Wu L, Kong B, et al. The First Nationwide Multicenter Prevalence Study of Germline BRCA1 and BRCA2 Mutations in Chinese Ovarian Cancer Patients. Int J Gynecol Cancer 2017;27:1650-7. [Crossref] [PubMed]
- Li W, Shao D, Li L, et al. Germline and somatic mutations of multi-gene panel in Chinese patients with epithelial ovarian cancer: a prospective cohort study. J Ovarian Res 2019;12:80. [Crossref] [PubMed]
- You Y, Li L, Lu J, et al. Germline and Somatic BRCA1/2 Mutations in 172 Chinese Women With Epithelial Ovarian Cancer. Front Oncol 2020;10:295. [Crossref] [PubMed]
- Hirotsu Y, Nakagomi H, Sakamoto I, et al. Multigene panel analysis identified germline mutations of DNA repair genes in breast and ovarian cancer. Mol Genet Genomic Med 2015;3:459-66. [Crossref] [PubMed]
- Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405-24. [Crossref] [PubMed]
- Boussios S, Karathanasi A, Cooke D, et al. PARP Inhibitors in Ovarian Cancer: The Route to "Ithaca". Diagnostics (Basel) 2019;9:55. [Crossref] [PubMed]
- AlHilli MM, Becker MA, Weroha SJ, et al. In vivo anti-tumor activity of the PARP inhibitor niraparib in homologous recombination deficient and proficient ovarian carcinoma. Gynecol Oncol 2016;143:379-88. [Crossref] [PubMed]
- Boussios S, Karihtala P, Moschetta M, et al. Combined Strategies with Poly (ADP-Ribose) Polymerase (PARP) Inhibitors for the Treatment of Ovarian Cancer: A Literature Review. Diagnostics (Basel) 2019;9:87. [Crossref] [PubMed]
- King MC, Marks JH, Mandell JB, et al. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science 2003;302:643-6. [Crossref] [PubMed]
- Pal T, Permuth-Wey J, Betts JA, et al. BRCA1 and BRCA2 mutations account for a large proportion of ovarian carcinoma cases. Cancer 2005;104:2807-16. [Crossref] [PubMed]
- Helpman L, Zidan O, Friedman E, et al. Young Israeli women with epithelial ovarian cancer: prevalence of BRCA mutations and clinical correlates. J Gynecol Oncol 2017;28:e61 [Crossref] [PubMed]
- Paradiso AV, Digennaro M, Patruno M, et al. BRCA germline mutation test for all woman with ovarian cancer? BMC Cancer 2019;19:641. [Crossref] [PubMed]
- Pennington KP, Walsh T, Harrell MI, et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin Cancer Res 2014;20:764-75. [Crossref] [PubMed]


