
Differences in the clinicopathological characteristics of pure and mixed invasive micropapillary breast carcinomas from eastern China
Introduction
Invasive micropapillary carcinoma of the breast (IMPC) is a rare pathologic subtype of breast cancer which accounts for 3–8% of all breast cancers (1-4). It is characterized by cohesive morula-like tumor cell clusters surrounded by clear empty spaces mimicking lymphatic vessels (5). It is well known that IMPC is associated with more frequent nodal metastasis, local recurrence, and distant metastasis compared with invasive ductal carcinoma of no special type (IDC-NST) (6,7). However, reports on the differences in outcome between IMPC and IDC-NST are inconclusive. Chen et al. reported that the 10-year survival rate of IMPC was significantly lower than that of IDC-NST (1), whereas other a majority of reports have found no significant prognostic difference for IMPC (8-11). IMPC is further classified as pure and mixed subtypes according to the degree of micropapillary differentiation and the proportion of IDC-NST component; as compared to pure IMPC, the mixed subtype is more common and has a major component of IDC (7,12). Mixed IMPC are more similar in clinical and genomic features to pure IMPC than to IDC-NSTs (13).
Investigations of the difference in pathological features between pure and mixed IMPC are scarce. Some retrospective studies have reported on the pathologic features of IMPC (6,7,12), but few have focused on the difference between the two subtypes. Whether pure IMPC can be used as an independent prognostic factor for breast cancer is still unknown. To the best of our knowledge, no investigation has been conducted on the difference in clinicopathological features between the pure and mixed IMPCs in a Chinese population. Therefore, we aimed to investigate the difference in clinicopathological characteristics [clinicopathological features, molecular subtype, primary tumor size, metastatic lymph node, 3-year disease-free survival (DFS), and overall survival (OS)] between localized pure and mixed IMPCs, using localized operable IMPC cases diagnosed and treated in our institute between 2010 and 2018.
We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-8045).
Methods
Patient population
We retrospectively recruited a total of 121 patients who were diagnosed with localized operable IMPCs, either pure or mixed type, and who were treated in Zhejiang Cancer Hospital, China, between May 2010 and August 2018. The enrollment criteria for the IMPC cases were selected strictly in accordance with the morphological criteria described in the WHO histological classification of breast tumors (14). The use of primary and metastatic tissue samples in this study was approved by the Ethics Committee of the Zhejiang Cancer Hospital. Written informed consent was acquired from all participants prior to the clinical data collection and tissue sample sequencing. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). Patients who had passed away were exempted from the informed consent but were also included in our analysis. Follow-up information was acquired from outpatient departmental records and additional personal telephone contact.
Clinicopathological characteristics
The detailed clinicopathological characteristics were as follows: age, histologic type, tumor size, TNM staging, Ki-67 index, number of metastatic lymph nodes, presence of blood vessel invasion, estrogen receptor (ER) status, progesterone receptor (PR) status, and HER2 overexpression and its subtypes based on immunohistochemical (IHC) classification [Luminal A, Luminal B (HER2 positive or negative), HER2 positive, and triple-negative]. The status of ER, PR, HER2 and Ki-67 was determined by IHC assay, which was a standard procedure performed by our pathology department staff. ER and PR were considered positive if 1% of the tumor cells were positive in nuclear staining. HER2 status was reported as positive with an IHC score of 3+ based on complete membranous staining in >10% of cells or with amplification confirmed by florescent in situ hybridization. According to the 2015 St Gallen consensus, five molecular subtypes of breast cancer have been identified: Luminal A (defined as ER positive, PR positive, Ki-67 30%, and HER2 negative), Luminal B & HER2+ (defined as ER positive, HER2 positive, regardless of PR status), HER2 overexpression (ER and PR negative, and HER2 positive), and triple negative (ER, PR, and HER2 negative).
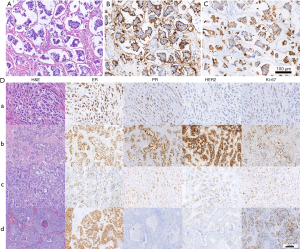
Slides stained with hematoxylin and eosin (H&E) from all blocks for each tumor were blindly and independently assessed and reviewed by two senior breast pathologists from our institute according to the morphologic criteria for the histologic classification of breast tumors from the World Health Organization (WHO). Differences of opinion were discussed until a consensus was reached. IMPC is characterized by “morule-like clusters” of tumor cells and surrounded by clear spaces with “inverted polarity” (Figure 1A). We defined the pure type as constituting at least 50% of the micropapillary component of the tumor. IHC staining of the epithelial membrane antigen (EMA) and mucin 1 (MUC1) were performed to certify the inverted polarity pattern (Figure 1B,C).
DFS was determined from the date of surgery to the date of relapse. The 3-year DFS rate was defined as the ratio of patients whose DFS was longer than 3 years. Patients whose follow-up duration was shorter than 3 years were excluded from the survival analyses. Overall survival (OS) was calculated from the date of surgery to the date of death.
Tissue DNA isolation and capture-based targeted DNA sequencing
DNA isolation and targeted sequencing were performed in Burning Rock Biotech, a clinical laboratory accredited by the College of American Pathologist (CAP) and certified by the Clinical Laboratory Improvement Amendments (CLIA), according to optimized protocols as described previously (15,16). Briefly, tissue DNA was extracted from the formalin-fixed, paraffin-embedded (FFPE) tumor tissues using the QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany). A minimum of 50 ng DNA was required for the next generation sequencing (NGS) library construction. Tissue DNA was sheared using M220 Focused Ultrasonicator (Covaris, MA, USA). Fragments between 200–400 bp were purified (Agencourt AMPure XP Kit, Beckman Coulter, CA, USA), hybridized with capture probes baits, selected with magnetic beads, and amplified. The quality and the size of the fragments were assessed by high sensitivity DNA kit using Bioanalyzer 2100 (Agilent Technologies, CA, USA). Target capture was performed using a commercial panel consisting of 520 genes (OncoScreen Plus, Burning Rock Biotech. Guangzhou, China), spanning 1.64 megabases of the human genome. Indexed samples were sequenced on Nextseq500 (Illumina, Inc., USA) with paired-end reads and an average sequencing depth of 1,000×.
Sequence data analysis
Sequence data were mapped to the reference human genome (hg19) using the Burrows-Wheeler Aligner v.0.7.10 which is a publicly available algorithm developed by Li and Durbin from Wellcome Trust Sanger Institute and published in Bioinformatics (17). Local alignment optimization and variant calling were performed using the Genome Analysis Tool Kit v.3.2, and VarScan v.2.4.3. Variants were filtered using the VarScan fpfilter pipeline, and loci with a depth less than 100 were filtered out. Base calling in tissue samples required at least 8 and 5 supporting reads for single nucleotide variations (SNV) and insertion-deletion variations (INDEL), respectively. Variants with a population frequency over 0.1% in the ExAC, 1000 Genomes, dbSNP, or ESP6500SI-V2 databases were grouped as single nucleotide polymorphisms (SNP) and were excluded from further analyses. The remaining variants were annotated with ANNOVAR and SnpEff v.3.6. Analyses of DNA translocation were performed using Factera v.1.4.3. Copy number variations (CNV) were analyzed on the basis of the depth of coverage data on capture intervals using an in-house algorithm. The limit of detection for CNVs was set at 1.5 and 2.64 for deletions and amplifications, respectively. Metastasis-specific genes refer to the genes that were not detected from the primary breast tissue, or where the allele fraction of the mutations was more than 1.2-fold elevated, compared to the primary tissue.
The tumor mutation burden (TMB) per patient was computed as a ratio of the total number of mutations divided by the total coding region size of the panel used (i.e., 1.64 Mb for the 520-gene OncoScreen Plus panel) using the formula below. CNVs, fusions, large genomic rearrangements, and mutations occurring on the kinase domain of EGFR and ALK were excluded from the mutation count.
[1]
To understand the somatic mutations that are shared and distinct for the primary breast tissues and metastatic tissues, phylogenetic trees were constructed for each of the 6 evaluable patients using Microsoft PowerPoint. Inferred evolution of the mutations shared between the primary and metastatic tissues formed the trunk, and the mutations not present in either the primary or metastatic tissues formed the branches. The length of the trunk and branches was scaled by the total number of mutations in the samples.
Statistical analysis
All statistical analyses were performed using R statistical software (R version 3.5.3; R Foundation for Statistical Computing; Vienna, Austria). Chi-squared and t-tests were performed to compare the clinicopathological differences enumerated in the table between the two groups in the study. Survival analyses of DFS and OS were evaluated by the Kaplan-Meier method, and the differences between the two groups were assessed using the log-rank test. A two-sided P value
Results
Patient characteristics
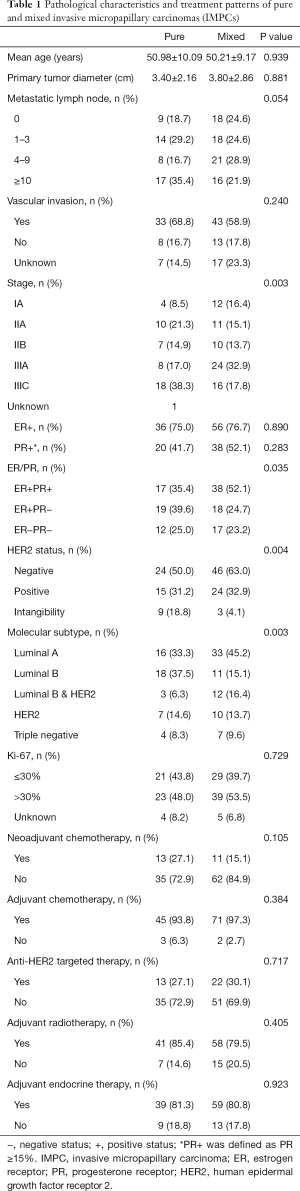
A total of 121 cases were identified and divided into two groups: pure IMPC (n=48) and mixed IMPC (n=73). A flow diagram was shown in Figure S1. The tumor size, molecular subtype, lymph node metastasis and 3-year disease free survival (DFS) rate of the two groups were compared. The pathological characteristics of the pure and mixed IMPCs are presented in Table 1. The mean age at diagnosis was 51.0±10.1 years for pure IMPC and 50.2±9.2 years for mixed IMPC (P=0.939). The mean diameter of the primary tumor was similar (3.4 cm for the pure IMPC and 3.8 cm for the mixed IMPC, P=0.881). No significant difference in lymph node metastasis, lymphovascular invasion, or the Ki-67 index was found between the two groups. A higher proportion of stage IIIc cases was observed in the pure group compared with the mixed group (38.3% vs. 17.8%; P=0.003).
Full table
No significant difference in ER/PR expression was observed between the two groups. In the pure group, the proportion of HER2 negative, HER2 positive, and HER2 intangibility was 50.0%, 31.2%, and 18.8%, respectively, whereas in the mixed group, the proportion was 63.0%, 32.9%, and 4.1%, respectively (P=0.004). The pure IMPC group had a lower proportion of Luminal A but a higher proportion of Luminal B compared with the mixed IMPC group (33.3% vs. 45.2%, and 37.5% vs. 15.1%, respectively; P=0.003). Representative images of Luminal A and Luminal B in the pure and mixed groups were shown in Figure 1D.
All patients received modified radical mastectomy or breast-conserving surgery. Axillary lymph node dissection was performed on patients who had positive node metastasis. Systemic chemotherapy was administered to patients by their attending physician according to the latest guidelines, including the regimens containing anthracycline, taxanes, or both. The amount of neoadjuvant or adjuvant chemotherapy did not differ significantly between the two groups, and all patients received adjuvant radiation therapy (RT) in accordance with the guidelines. Adjuvant endocrine therapy was administered to 39 patients (81.3%) with pure IMPC and 59 patients (80.8%) with mixed IMPC who were positive for hormone receptors. Anti-HER2 targeted therapy was used for patients who had an overexpression of HER2. Of these patients, 13 (27.1%) were pure IMPC and 22 (30.1%) were mixed IMPC. However, there was no difference between the two groups.
Comparison of survival outcomes
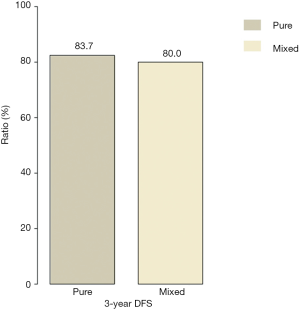
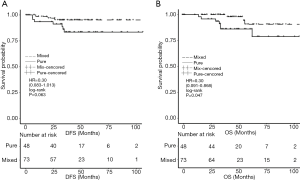
Follow-up information was available for 113 cases after excluding eight patients whose follow-up duration was shorter than 3 years. The median follow-up time was 48.9 months. The 3-year DFS rates were 83.7% (36/43) and 80.0% (56/70) in the pure and mixed IMPC groups, respectively (P=0.772) (Figure 2). The analysis of DFS between the two groups showed a marginally significant difference [HR =0.30 (0.083–1.013), P=0.063]. However, the mixed group had a significantly higher OS rate compared with the pure IMPC group [HR =0.28 (0.091–0.868), P=0.047] (Figure 3). Ten deaths were identified (seven with pure IMPC and three with mixed IMPC) constituting an overall mortality of 8.8%.
Molecular features of pure and mixed type IMPC
Targeted sequencing was performed to interrogate the genomic profile on paired primary breast cancer and metastatic tissue samples from two patients with pure type IMPC and four patients with mixed type IMPC (SRA accession: PRJNA594098, https://www.ncbi.nlm.nih.gov/sra/PRJNA594098). The molecular subtypes of the two pure IMPC patients were Luminal B and triple negative breast cancer. Among the four mixed IMPC patients, two were triple negative, one was Luminal B & HER2+ and one was HER2 overexpression breast cancer. For the pure group, a total of 25 mutations in 23 genes from the primary tissue and 32 mutations in 28 genes from the metastatic tissue were detected, while for the mixed group, a total of 26 mutations in 19 genes from the primary tissue and 29 mutations in 22 genes from the metastatic tissue were detected. TP53 mutations were detected in the primary and metastatic tissues from all six patients. In addition, PIK3CA mutations were detected in the primary and metastatic tissues from four patients with mixed type and from one patient with pure type. KRAS G12A [allelic fraction (AF) =17.86%] and an increase in allelic fraction of KRAS G12A (AF =73.28%) jointly with KRAS copy number amplification (copy number =6.51) were detected in the primary and metastatic tissues from one patient with pure type. BRCA Q147R was detected in the primary and metastatic tissues from one patient, while BRCA T855A was detected only in the primary tissue from another patient (Figure 4A).
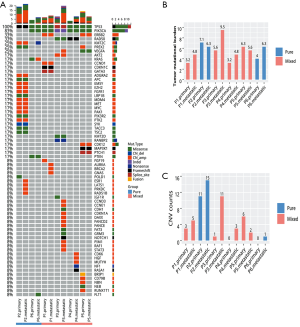
The two pure type patients had an average TMB of 5.55 mutations/Mb from the primary tissues and 6.3 mutations/Mb from the metastatic tissues. In contrast, the four patients with mixed type had an average TMB of 4.58 mutations/Mb from the primary tissues and 6.18 mutations/Mb from the metastatic tissues (Figure 4B).
Interestingly, 13 mutations in 12 genes and 31 mutations in 28 genes were found to be metastatic-specific for the pure type and mixed type patients, respectively. Among these mutations, a total of 16 CNVs in 16 genes were only detected in the metastatic tissues from three patients with mixed IMPC (Figure 4C). Pathway analysis revealed that the PI3K-AKT signaling pathway was the most significantly mutated pathway in the metastatic tissues.
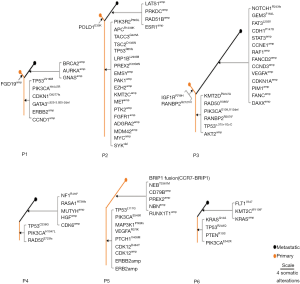
The phylogenetic trees are presented in Figure 4 (for two patients with pure IMPC and four patients with mixed IMPC). For patients P1, P2 and P3, the metastatic tissue had more specific genes compared to the primary tissue, although both tissues had specific genes. Metastatic-specific mutations were found only for patients P4 and P6, while primary-specific mutation genes were only found for patient P5 (Figure 5).
Discussion
We systematically investigated the difference in clinicopathological characteristics between pure and mixed IMPC in patients from eastern China. We found that pure IMPCs had a higher proportion of stage IIIc cases compared to the mixed IMPCs, which suggests pure IMPC has a more aggressive behavior. This finding is also in line with a previous study reporting a higher incidence of lymph node metastasis at initial diagnosis for IMPC compared to IDC-NST (1). Similar to previous studies, we also found that pure IMPCs have a higher proportion of lymph node metastasis (81.3% vs. 75.4%) and N3 (more than nine lymph node metastasis; 35.4% vs. 21.9%), compared with mixed IMPCs. ER and PR profiles between the two groups were similar. Notably, we also found that pure IMPCs had a higher rate of the Luminal B subtype than the mixed group, while the mixed IMPCs were more likely to be Luminal A (P=0.003). Taken together, our findings suggest that pure IMPCs has a more aggressive behavior compared with mixed IMPCs.
We found that the mixed IMPC group had a longer overall survival compared with the pure group, but in contrast to other studies (18), no significant difference in DFS or the 3-year DFS rate was found between the two groups (83.7% vs. 80.0%). The mixed group showed a trend towards a better prognosis (DFS) compared to the pure group [HR =0.30 (0.08–1.01)], but this difference did not reach statistical significance (P=0.063). This result may be due to the short follow-up duration and small sample size. Additionally, the short follow-up and low incidence of deaths in our study might preclude a definitive conclusion that pure and mixed IMPCs have a different clinical prognosis. Besides, the outcomes between IMPC and IDC-NST have controversial conclusions among the studies. Because precise treatment based on the molecular classification of breast cancer results in different outcomes between different molecular subtypes, to a certain extent, the difference in prognosis between IMPC and IDC-NST is difficult to analyze simply from the pathological type. So whether there is prognostic difference for IMPC or not needs a well-designed prospective research to confirm.
Growing efforts are exerted to understand the molecular features of IMPC; however, due to its rarity, the molecular mechanisms on the difference in pure and mixed IMPC remain unclear. Many studies have reported that IMPC has a different molecular phenotype from IDC-NST. Comprehensive investigation on small non-coding RNA transcriptome identified a number of differentially expressed microRNA (miRNA) involved in the regulation of biological processes including let-7b, miR-30c, miR-148a, miR-181a, and miR-181b in IMPCs compared with IDC-NST (19). Another study showed that the gains of 1q and the losses of 6q, 17p, 19p, and 22q were more prevalent in IMPC than in IDC-NST (20). In addition, the differential expression of prostate stem cell antigen (PSCA), LZTS1, plakoglobin, MUC1, and MUC4 were reported to be associated with IMPC (21-24). The aforementioned molecular characteristics are related to the high frequency of lymph vessel invasions (LVIs) and lymph node metastases (LNMs,), the maintenance of cell morphology, and the high metastatic propensity in IMPC (21-24). Pangenomic analysis stratified IMPCs into two molecular subsets: one subset is characterized by the increased 16p amplifications (71%), while the other subset is characterized by increased amplifications of 8q (35%), 17q (20–46%), and 20q (23–30%) and loss of 17p (74%) (25). Despite the lack of canonical driver mutations, gene ontology analysis revealed that IMPCs were enriched with various somatic mutations in genes involved in cellular polarity (i.e., DNAH9 and FMN2), ciliogenesis (i.e., BBS12 and BBS9), endoplasmic reticulum-related proteins (i.e., HSP90B1 and SPTLC3), and cytoskeleton (i.e., UBR4 and PTPN21), indicating their contribution to the distinct morphological features of IMPCs (25). Similar to common breast cancer histology, PIK3CA, TP53, GATA3, and MAP2K4 have been the most commonly mutated genes among 29 IMPCs (26). Marchiò et al. also reported that cyclin D1 overexpression and MYC (8q24) amplification were associated with pure IMPCs (27). Furthermore, in another study, Marchiò et al. demonstrated that mixed IMPCs have a similar genetic and IHC profile to pure IMPCs rather than IDC-NSTs (13). In our study, we found FGFR1, a tyrosine kinase receptor known to be related to the survival of cancer cells, was amplified only in one pure IMPC patient, while Marchiò et al. showed that overexpression of FGFR1 existed in four out of 12 mixed IMPCs (13). This discrepancy in findings may result from the limited sample size in our study, and further analysis of genetic profiling is warranted. Consistent with the study by Dieci et al. (26), the most frequently mutated genes in our cohort, were TP53 and PIK3CA, which were detected in both primary breast tumors and metastatic tissues.
Additionally, we compared the genomic alterations between the primary and metastatic tumors. The result showed that the new-onset genetic alterations of the metastatic tumors were the overexpression of BRCA2, ESR1, STAT3, VEGFA, CDK6 and CCND3 and the mutation of NOTCH1, CDH1, and NEB, etc. Pathway analysis revealed that the PI3K-AKT signaling pathway was the most significantly mutated pathway in the metastatic tumors, which may potentially offer additional therapeutic opportunities. In addition, the different profiles of the phylogenetic trees of six patients reflected the tumor heterogeneity among different individuals. Further functional studies investigating driver mutation or amplicon sequencing for pure and mixed IMPCs are highly warranted.
Our study had a number of strengths and limitations. This is the first study from China systematically investigating the difference in clinicopathological features between pure and mixed IMPC and comparing the genomic profiles from primary and metastatic tumors. We found the mixed group had a statistically significant higher OS compared to the pure group. However, our small sample size and short follow-up time limited the analysis of DFS, and we did not investigate the prognostic factors between the two subtypes.
In conclusion, we found that pure IMPC had a more aggressive behavior with locally advanced disease and a higher proportion of Luminal B than mixed IMPC. Mixed IMPC had a longer OS compared to pure IMPC, but there was no significant difference in 3-year DFS rate between the two groups. A larger sample size and a longer clinical follow-up are necessary to validate our findings. Further investigation of the molecular mechanism differences between the two groups is highly warranted.
Acknowledgments
We thank Tianhui Chen and Hong Zhang for their modifications to this paper. We also acknowledge Yongran Cheng for his excellent assistance in statistical analysis.
Funding: This work was funded by grants from the National Natural Science Foundation of China (grant number: 81672597), the Key Research-Development Program of Zhejiang Province (grant number: 2017C03013, 2019C04001), the Natural Science Foundation of Zhejiang Province, China (grant number: LY17H160038, LY21H160005), the Joint Key Program of Zhejiang Province- Ministry of Health (grant number: WKJ-ZJ-1714), and the Qianjiang Talents Fund of Zhejiang Province (grant number: QJD1602026).
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-8045
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-8045
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-8045). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The use of primary and metastatic tissue samples in this study was approved by the Ethics Committee of the Zhejiang Cancer Hospital. Written informed consent was acquired from all participants prior to the clinical data collection and tissue sample sequencing. Patients who had passed away were exempted from the informed consent but were also included in our analysis. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen L, Fan Y, Lang RG, et al. Breast carcinoma with micropapillary features: clinicopathologic study and long-term follow-up of 100 cases. Int J Surg Pathol 2008;16:155-63. [Crossref] [PubMed]
- Chen AC, Paulino AC, Schwartz MR, et al. Prognostic markers for invasive micropapillary carcinoma of the breast: a population-based analysis. Clin Breast Cancer 2013;13:133-9. [Crossref] [PubMed]
- Guo X, Chen L, Lang R, et al. Invasive micropapillary carcinoma of the breast: association of pathologic features with lymph node metastasis. Am J Clin Pathol 2006;126:740-6. [Crossref] [PubMed]
- Ide Y, Horii R, Osako T, et al. Clinicopathological significance of invasive micropapillary carcinoma component in invasive breast carcinoma. Pathol Int 2011;61:731-6. [Crossref] [PubMed]
- Siriaunkgul S, Tavassoli FA. Invasive micropapillary carcinoma of the breast. Mod Pathol 1993;6:660-2. [PubMed]
- Zekioglu O, Erhan Y, Ciris M, et al. Invasive micropapillary carcinoma of the breast: high incidence of lymph node metastasis with extranodal extension and its immunohistochemical profile compared with invasive ductal carcinoma. Histopathology 2004;44:18-23. [Crossref] [PubMed]
- Hasan Gokce MD, Durak MG, Akin MM, et al. Invasive Micropapillary Carcinoma of the Breast: A Clinicopathologic Study of 103 Cases of an Unusual and Highly Aggressive Variant of Breast Carcinoma. Breast Journal 2013;19:374-81. [Crossref] [PubMed]
- Gokce H, Durak MG, Akin MM, et al. Invasive micropapillary carcinoma of the breast: a clinicopathologic study of 103 cases of an unusual and highly aggressive variant of breast carcinoma. Breast J 2013;19:374-81. [Crossref] [PubMed]
- Nassar H, Wallis T, Andea A, et al. Clinicopathologic analysis of invasive micropapillary differentiation in breast carcinoma. Mod Pathol 2001;14:836-41. [Crossref] [PubMed]
- Hao S, Zhao YY, Peng JJ, et al. Invasive micropapillary carcinoma of the breast had no difference in prognosis compared with invasive ductal carcinoma: a propensity-matched analysis. Sci Rep 2019;9:286. [Crossref] [PubMed]
- Vingiani A, Maisonneuve P, Dell'orto P, et al. The clinical relevance of micropapillary carcinoma of the breast: a case-control study. Histopathology 2013;63:217-24. [Crossref] [PubMed]
- Shi WB, Yang LJ, Hu X, et al. Clinico-pathological features and prognosis of invasive micropapillary carcinoma compared to invasive ductal carcinoma: a population-based study from China. PLoS One 2014;9:e101390. [Crossref] [PubMed]
- Marchiò C, Iravani M, Natrajan R, et al. Mixed micropapillary-ductal carcinomas of the breast: a genomic and immunohistochemical analysis of morphologically distinct components. J Pathol 2009;218:301-15. [Crossref] [PubMed]
- Lakhani S EI, Schnitt S et al. WHO classification of tumours of the breast, 4th edn. IARC Press, Lyon 2012.
- Tao Z, Li T, Feng Z, et al. Characterizations of Cancer Gene Mutations in Chinese Metastatic Breast Cancer Patients. Front Oncol 2020;10:1023. [Crossref] [PubMed]
- Chen B, Zhang G, Li X, et al. Comparison of BRCA versus non-BRCA germline mutations and associated somatic mutation profiles in patients with unselected breast cancer. Aging (Albany NY) 2020;12:3140-55. [Crossref] [PubMed]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754-60. [Crossref] [PubMed]
- Kaya C, Ucak R, Bozkurt E, et al. The Impact of Micropapillary Component Ratio on the Prognosis of Patients With Invasive Micropapillary Breast Carcinoma. J Invest Surg 2020;33:31-9. [Crossref] [PubMed]
- Li S, Yang C, Zhai L, et al. Deep sequencing reveals small RNA characterization of invasive micropapillary carcinomas of the breast. Breast Cancer Res Treat 2012;136:77-87. [Crossref] [PubMed]
- Natrajan R, Wilkerson PM, Marchio C, et al. Characterization of the genomic features and expressed fusion genes in micropapillary carcinomas of the breast. J Pathol 2014;232:553-65. [Crossref] [PubMed]
- Hao JY, Yang YL, Li S, et al. PSCA expression in invasive micropapillary carcinoma of breast. Zhonghua Bing Li Xue Za Zhi 2011;40:382-6. [PubMed]
- Wang XX, Liu BB, Wu X, et al. Loss of Leucine Zipper Putative Tumor Suppressor 1 (LZTS1) Expression Contributes to Lymph Node Metastasis of Breast Invasive Micropapillary Carcinoma. Pathol Oncol Res 2015;21:1021-6. [Crossref] [PubMed]
- Nassar H, Pansare V, Zhang H, et al. Pathogenesis of invasive micropapillary carcinoma: role of MUC1 glycoprotein. Mod Pathol 2004;17:1045-50. [Crossref] [PubMed]
- Huang L, Ji H, Yin L, et al. High Expression of Plakoglobin Promotes Metastasis in Invasive Micropapillary Carcinoma of the Breast via Tumor Cluster Formation. J Cancer 2019;10:2800-10. [Crossref] [PubMed]
- Gruel N, Benhamo V, Bhalshankar J, et al. Polarity gene alterations in pure invasive micropapillary carcinomas of the breast. Breast Cancer Res 2014;16:R46. [Crossref] [PubMed]
- Dieci MV, Smutna V, Scott V, et al. Whole exome sequencing of rare aggressive breast cancer histologies. Breast Cancer Res Treat 2016;156:21-32. [Crossref] [PubMed]
- Marchiò C, Iravani M, Natrajan R, et al. Genomic and immunophenotypical characterization of pure micropapillary carcinomas of the breast. J Pathol 2008;215:398-410. [Crossref] [PubMed]


