
Evaluating the association between single nucleotide polymorphisms in the stonin 2 (STON2) gene and keratoconus in a Han Chinese population
Introduction
Keratoconus (KCTN) is a major indication for cornea transplantation in developed countries (1). It is characterized by progressive corneal thinning and conical protrusion, leading to severe visual disturbances (2). KCTN has an average prevalence of 1:2,000 globally but varies across different ethnicities. Asians are reported to have a remarkably higher incidence of KCTN than Caucasians, suggesting that ethnic differences might have considerable impacts on this disease (3). The clinical management of KCTN varies depending on the clinical stages. In the early stages, collagen UV cross-linking or contact lenses can stabilize the cornea biomechanics, thereby controlling KCTN progression (4). Therefore, early diagnosis of KCTN based on a clear understanding of its etiology may help guide clinical interventions.
The etiology of KCTN is multifactorial, combining both genetic and non-genetic factors. Environmental factors including UV exposure (5), contact lens wear (6), atopy (7), and constant eye rubbing (8) can all act as triggers of the condition in genetically predisposed individuals. However, more importantly, a strong genetic susceptibility underlying KCTN pathogenesis has been clearly demonstrated in familial aggregation studies (9,10), twins studies (11), linkage analyses, and genome-wide association studies (GWASs) (4,12). So far, over 150 single polynucleotide polymorphisms (SNPs) in more than 60 loci have been identified to be linked with KCTN susceptibility (13-20). Some SNPs were discovered in GWASs examining central corneal thickness (CCT), which is a critical quantitative trait locus (QTL) associated with increased KCTN risk. These identified loci include MDPZ-NF1B, FOXO1, FND3B, COL4A3, COL4A4, and COL5A1 (21-23). While many of these reported loci have been detected in several other ethnicities, inconsistent results exist across different cohorts, rendering their contribution to KCTN risk inconclusive (18,24-27).
Recent GWASs identified the stonin 2 gene (STON2) as a susceptibility gene for CCT (21,23). STON2 encodes a membrane protein that is involved in regulating endocytotic complexes. It is one of the clathrin-associated adaptor molecules which ensures that specific proteins are internalized (28). Hosoda et al. (23) discovered that the STON2 allele rs2371597, which was associated with increased CCT, was also associated with an increased risk of KCTN in the Japanese population. However, due to the underlying genetic heterogeneity of KCTN susceptibility, it remains unknown whether this SNP also confers the risk of KCTN susceptibility in other cohorts. It is also possible that other additional SNPs located in the same locus might contribute to KCTN susceptibility. Therefore, an extensive investigation on this locus is warranted. Herein, a replication study on the reported SNP rs2371597, together with two potential regulatory SNPs within the same locus, was carried out to explore the contribution of the STON2 locus to KCTN susceptibility in an independent Han Chinese population.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-6654).
Methods
Subjects
A total of 164 Han Chinese patients with KCTN were randomly selected from the Department of Ophthalmology at the Eye and ENT Hospital of Fudan University between October 2015 and November 2018. All patients were self-reported as Han Chinese ethnicity. Patients were diagnosed with KCTN if they presented with at least one KCTN symptom, such as corneal stromal thinning, Fleischer’s ring, Vogt’s striae, or Munson’s sign, and videokeratography showed refractive errors or signs. Patients with KCTN and syndromic disease (such as Down’s syndrome and Leber optic atrophy) were excluded. A total of 239 healthy individuals with no signs of ocular disorder were included as the control group.
The study was conducted following the Declaration of Helsinki (as revised in 2013) and has been approved by the Ethics Committee of the Eye and ENT Hospital of Fudan University (2015044-1). Informed consent was obtained from all participants.
Genomic DNA extraction
Genomic DNA was extracted from the peripheral blood monocytes using the QIAGEN FlexiGene DNA kit (Qiagen, Hamburg, Germany) following the manufacturer’s protocol. DNA concentration was measured with a NanoDrop spectrophotometer (NanoDrop Technologies, DE, USA).
Genotyping of SNPs
Three SNPs (rs2371597, rs8004137, and rs8008602) located in the STON2 gene were selected for investigation, utilizing the TaqMan SNP Genotyping Assays (C__2791741_10 for rs2371597, C__29283383_10 for rs8004137, and C__29283385_10 for rs8008602). The genotyping assay was performed using real-time polymerase chain reaction (PCR; Applied Biosystems VII, USA) with the standard genotyping protocol. Each reaction contained 2 μL DNA template, 5 μL of MasterMix buffer (2× concentration; ThermoFisher), 2.5 μL double distilled water (ddH2O), and 0.25 μL probe (40× concentration). Genotypes were automatically determined using the ratio of VIC and FAM fluorescent signals from the TaqMan assay.
Data analysis
Data analysis was conducted with the PLINK package (29). SNP frequency in case and control group was tested for departure from HWE with an exact test. A χ2-test was used to calculate the difference in SNP allele frequency between cases and controls. The logistic regression model, adjusted by gender and age, was used to calculate the odds ratios (ORs) and the 95% confidence intervals (CIs). P values less than 0.05 were considered statistically significant. The LDlink package was used to calculate the linkage disequilibrium (LD) within SNPs (30). The RegulomeDB (31) tool was applied to annotate SNPs with known or predicted regulatory elements. Meta-analysis was conducted by weighting effect size estimates with the inverse of the standard errors. The SNP association’s statistical significance was calculated using the Z-test, and a pooled P value <0.05 was considered significant.
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
Male Balb/c mice (6–8 weeks old) were obtained from Shanghai Laboratory Animal Center, Chinese Academy of Sciences (CAS). Mice were anesthetized by pentobarbital sodium and perfused by intracardiac injection of phosphate-buffered saline (PBS). Murine organ tissues, including the cornea, retina, heart, liver, lung, kidney, brain, and lymph nodes, were isolated. Total RNA was extracted with RNA Simple Total RNA Kit (TIANGEN, Beijing, China). The first-strand cDNA was reverse transcribed with the FastKing RT Kit (TIANGEN, Beijing, China). Quantitative real-time PCR was performed with a QuantiNova SYBR Green PCR Kit (Qiagen, Hamburg, Germany) according to the manufacturer’s protocols. Primer sequences used are listed below.
Mouse glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene:
Forward 5'-CATCACTGCCACCCAGAAGACTG-3',
Reverse 5'-ATGCCAGTGAGCTTCCCGTTCAG-3';
Mouse STON2 gene:
Forward 5'-GTATGAGCACGCCTTCAACTCC-3',
Reverse 5'- GGCAAATCTGGAAGGCACTTCC-3'.
All reactions were performed in quadruplicate, and the experiments were replicated three times independently. Relative gene expression levels were determined using the delta Ct method. Animal experiments were performed under a project license (2015044-2) granted by the Ethics Committee of the Eye and ENT Hospital of Fudan University, in compliance with the Association for Research in Vision and Ophthalmology (ARVO) statement for the Use of Animals in Ophthalmic and Vision Research.
Results
A total of 164 patients with KCTN and 239 healthy subjects were recruited in this study. The average age of patients with KCTN was 23.64±6.17 years, and 75.6% of this cohort were male (Table 1). The healthy controls had an average age of 30.02±5.36 years, and 62.7% were male. These characteristics were comparable to that of the KCTN group.

Full table
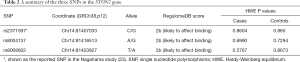
Three SNPs in the STON2 gene were genotyped. SNP rs2371597 has been shown in a GWAS to affect KCTN susceptibility in the Japanese population (23). The current study aimed to determine whether it had a similar effect on the Han Chinese population. The other two SNPs were selected based on in-silico functional annotation. Briefly, SNPs showing LD (r2≥0.8 in East Asian cohort from 1000 Genomes Project Phase 3) with rs2371597 were subjected to RegulomeDB annotation to examine if they overlapped with some known or predicted regulatory elements. Consequently, two other SNPs, namely rs8004137 and rs8008602, were selected for replication, given their predicted roles in affecting transcription factor binding (Table 2, Figure 1A).
Full table
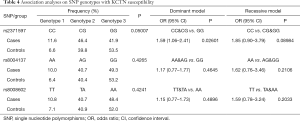
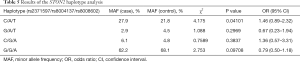
The average genotyping call rate was 95.0% for the three genotyped SNPs, and all satisfied the Hardy-Weinberg equilibrium (HWE) in the controls (Table 2 and Table S1). The association result showed that only the C allele of SNP rs2371597 was significantly associated with KCTN susceptibility (OR =1.47, 95% CI: 1.08 to 2.02, P=0.01409, P=0.04227 with Bonferroni correction, Table 3). A meta-analysis on the association results of the Japanese cohort (23) and this Chinese cohort further supported the association of rs2371597 with KCTN risk (P_meta =0.005694, OR_meta =1.37, 95% CI: 1.11 to 1.62). Genotype association analysis showed that, within the dominant model, genotype CC and CG of rs2371597 revealed an OR of 1.59 (95% CI: 1.06 to 2.41, P=0.02601, Table 4). The association results between the STON2 gene haplotype and the risk of KCTN are listed in Table 5. The haplotype CAT was significantly associated with the risk of KCTN (P=0.04101). The other two SNPs allele and genotypic frequencies did not reveal substantial differences between the cases and the controls.

Full table
Full table
Full table
Furthermore, gene expression pattern analysis among different murine organ tissues showed that STON2 was widely expressed in all mouse tissues examined and presented a relatively higher expression in the cornea (Figure 1B).
Discussion
This study demonstrated the association of the STON2 locus with KCTN susceptibility in a Han Chinese population. This concurs with a previous study in a Japanese population (23), and meta-analysis data supported this observation (P_meta 0.005694, OR_meta =1.37, 95% CI: 1.11 to 1.62). These results support the notion that STON2 contributes to KCTN susceptibility among Asians.
Segregation analyses have suggested that KCTN is a complicated trait affected by many factors, including genes, variable penetrance, and environmental triggers (32). Although some efforts have been made (4) to explore the genetic factors contributing to KCTN risk, the understanding of KCTN genetics is still in its infancy. In contrast to other complex diseases that usually recruit over thousands of participants, the sample sizes in GWASs examining KCTN have generally been quite small, thus limiting the statistical power for detecting novel association signals. Alternatively, the involvement of the endophenotype CCT is efficient in identifying additional susceptibility genes for KCTN (21). CCT is a highly heritable quantitative trait, with heritability estimates ranging from 0.68 to 0.95 (33,34). Thinner CCT is often observed in common ocular diseases such as KCTN (35). With such an approach, several recent GWASs have successfully identified that some of these CCT loci were also associated with KCTN susceptibility. STON2 was one such gene, and therefore it was the focus of this current study. The successful validation of the association between STON2 SNPs and KCTN risk in both the Japanese and the Han Chinese populations supports future investigations into other newly defined CCT loci and their association with KCTN susceptibility (21).
The current study demonstrated the association between STON2 and KCTN susceptibility among Asians. Interestingly, the effect size of SNP rs2371597 differed between the Chinese and the Japanese cohorts (OR =1.47 in the Chinese cohort, and OR =1.27 in the Japanese cohort), which may be partially due to the different minor allele frequencies between the two cohorts (C =32.5% in the Chinese cohort and C =25.5% in the Japanese cohort). Thus, it would be of interest to further examine the contribution of STON2 SNP(s) to KCTN susceptibility in other ethnicities, such as the Caucasian population. Furthermore, three SNPs with predicted roles in affecting binding were genotyped. However, two of them did not show any substantial association with KCTN in this cohort, although some trends were detected. Herein, in-depth replication studies with larger sample size are warranted. Lastly, the relationship between genetic variants and cellular abnormalities is still unclear. A previous Japanese study showed strong expression of STON2 in basal cells compared to superficial cells in the corneal epithelium, with minimal expression in the corneal stroma and endothelium layer. Our findings also highlighted a relatively high expression of STON2 in the cornea compared to other organ tissues. Deficiencies in STON2 may increase cell vulnerability to physical damage or immunological changes, thereby affecting KCTN development via interacting with extracellular matrix modeling. However, direct biological evidence will be required to confirm this hypothesis. Future systematically functional investigations should be conducted to elucidate further the mechanisms by which susceptible variants affect STON2 gene function and finally contribute to KCTN development.
In conclusion, this study explored the association between SNPs in the STON2 gene and KCTN susceptibility in a Han Chinese population. The results verified that the STON2 gene contributes to KCTN susceptibility. This study provided novel insights related to the genetic basis underlying KCTN pathogenesis, which will pave the way for future advances in prevention and treatment.
Acknowledgments
Funding: The authors were sponsored by the National Natural Science Foundation of China (81870630, 81700806). The sponsor or funding organization had no role in the design or conduct of this research.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-6654
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-6654
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-6654). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of the Eye and ENT Hospital of Fudan University (2015044-1) and informed consent was obtained from all participants. Animal experiments were performed under a project license (2015044-2) granted by the Ethics Committee of the Eye and ENT Hospital of Fudan University, in compliance with the ARVO statement for the Use of Animals in Ophthalmic and Vision Research.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Faria-Correia F, Luz A, Ambrosio R. Managing corneal ectasia prior to keratoplasty. Expert Rev Ophthalmo 2015;10:33-48. [Crossref]
- Rabinowitz YS. Keratoconus. Surv Ophthalmol 1998;42:297-319. [Crossref] [PubMed]
- Kok YO, Tan GF, Loon SC. Review: keratoconus in Asia. Cornea 2012;31:581-93. [Crossref] [PubMed]
- Karolak JA, Gajecka M. Genomic strategies to understand causes of keratoconus. Mol Genet Genomics 2017;292:251-69. [Crossref] [PubMed]
- Arnal E, Peris-Martinez C, Menezo JL, et al. Oxidative stress in keratoconus? Invest Ophthalmol Vis Sci 2011;52:8592-7. [Crossref] [PubMed]
- Steahly LP. Keratoconus following contact lens wear. Ann Ophthalmol 1978;10:1177-9. [PubMed]
- Bawazeer AM, Hodge WG, Lorimer B. Atopy and keratoconus: a multivariate analysis. Br J Ophthalmol 2000;84:834-6. [Crossref] [PubMed]
- McMonnies CW. Mechanisms of rubbing-related corneal trauma in keratoconus. Cornea 2009;28:607-15. [Crossref] [PubMed]
- Burdon KP, Vincent AL. Insights into keratoconus from a genetic perspective. Clin Exp Optom 2013;96:146-54. [Crossref] [PubMed]
- Naderan M, Rajabi MT, Zarrinbakhsh P, et al. Association between Family History and Keratoconus Severity. Curr Eye Res 2016;41:1414-8. [Crossref] [PubMed]
- Tuft SJ, Hassan H, George S, et al. Keratoconus in 18 pairs of twins. Acta Ophthalmol 2012;90:e482-6. [Crossref] [PubMed]
- Mackey DA, Hewitt AW. Genome-wide association study success in ophthalmology. Curr Opin Ophthalmol 2014;25:386-93. [Crossref] [PubMed]
- Li X, Bykhovskaya Y, Tang YG, et al. An association between the calpastatin (CAST) gene and keratoconus. Cornea 2013;32:696-701. [Crossref] [PubMed]
- Li X, Bykhovskaya Y, Canedo AL, et al. Genetic association of COL5A1 variants in keratoconus patients suggests a complex connection between corneal thinning and keratoconus. Invest Ophthalmol Vis Sci 2013;54:2696-704. [Crossref] [PubMed]
- Li X, Bykhovskaya Y, Haritunians T, et al. A genome-wide association study identifies a potential novel gene locus for keratoconus, one of the commonest causes for corneal transplantation in developed countries. Hum Mol Genet 2012;21:421-9. [Crossref] [PubMed]
- Cuellar-Partida G, Springelkamp H, Lucas SE, et al. WNT10A exonic variant increases the risk of keratoconus by decreasing corneal thickness. Hum Mol Genet 2015;24:5060-8. [Crossref] [PubMed]
- De Bonis P, Laborante A, Pizzicoli C, et al. Mutational screening of VSX1, SPARC, SOD1, LOX, and TIMP3 in keratoconus. Mol Vis 2011;17:2482-94. [PubMed]
- Wang Y, Jin T, Zhang X, et al. Common single nucleotide polymorphisms and keratoconus in the Han Chinese population. Ophthalmic Genet 2013;34:160-6. [Crossref] [PubMed]
- Czugala M, Karolak JA, Nowak DM, et al. Novel mutation and three other sequence variants segregating with phenotype at keratoconus 13q32 susceptibility locus. Eur J Hum Genet 2012;20:389-97. [Crossref] [PubMed]
- Bykhovskaya Y, Li X, Epifantseva I, et al. Variation in the lysyl oxidase (LOX) gene is associated with keratoconus in family-based and case-control studies. Invest Ophthalmol Vis Sci 2012;53:4152-7. [Crossref] [PubMed]
- Iglesias AI, Mishra A, Vitart V, et al. Cross-ancestry genome-wide association analysis of corneal thickness strengthens link between complex and Mendelian eye diseases. Nat Commun 2018;9:1864. [Crossref] [PubMed]
- Lu Y, Vitart V, Burdon KP, et al. Genome-wide association analyses identify multiple loci associated with central corneal thickness and keratoconus. Nat Genet 2013;45:155-63. [Crossref] [PubMed]
- Hosoda Y, Miyake M, Meguro A, et al. Keratoconus-susceptibility gene identification by corneal thickness genome-wide association study and artificial intelligence IBM Watson. Commun Biol 2020;3:410. [Crossref] [PubMed]
- Hao XD, Chen P, Chen ZL, et al. Evaluating the Association between Keratoconus and Reported Genetic Loci in a Han Chinese Population. Ophthalmic Genet 2015;36:132-6. [Crossref] [PubMed]
- Wang Y, Wei W, Zhang C, et al. Association of Interleukin-1 Gene Single Nucleotide Polymorphisms with Keratoconus in Chinese Han Population. Curr Eye Res 2016;41:630-5. [PubMed]
- Wang YM, Ma L, Lu SY, et al. Analysis of multiple genetic loci reveals MPDZ-NF1B rs1324183 as a putative genetic marker for keratoconus. Br J Ophthalmol 2018;102:1736-41. [Crossref] [PubMed]
- Zhang J, Wu D, Li Y, et al. Evaluating the association between calpastatin (CAST) gene and keratoconus in the Han Chinese population. Gene 2018;653:10-3. [Crossref] [PubMed]
- Martina JA, Bonangelino CJ, Aguilar RC, et al. Stonin 2: an adaptor-like protein that interacts with components of the endocytic machinery. J Cell Biol 2001;153:1111-20. [Crossref] [PubMed]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559-75. [Crossref] [PubMed]
- Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 2015;31:3555-7. [Crossref] [PubMed]
- Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 2012;22:1790-7. [Crossref] [PubMed]
- Kriszt A, Losonczy G, Berta A, et al. Segregation analysis suggests that keratoconus is a complex non-mendelian disease. Acta Ophthalmol 2014;92:e562-8. [Crossref] [PubMed]
- Dimasi DP, Burdon KP, Craig JE. The genetics of central corneal thickness. Br J Ophthalmol 2010;94:971-6. [Crossref] [PubMed]
- Toh T, Liew SH, MacKinnon JR, et al. Central corneal thickness is highly heritable: the twin eye studies. Invest Ophthalmol Vis Sci 2005;46:3718-22. [Crossref] [PubMed]
- Steele TM, Fabinyi DC, Couper TA, et al. Prevalence of Orbscan II corneal abnormalities in relatives of patients with keratoconus. Clin Exp Ophthalmol 2008;36:824-30. [Crossref] [PubMed]


