
UAP1L1 plays an oncogene-like role in glioma through promoting proliferation and inhibiting apoptosis
Introduction
Glioma is the most prevalent primary tumor of the brain and spinal cord. According to the World Health Organization (WHO) grading criteria, glioma is histologically categorized into grade I–IV subtypes (1). The invasion ability of glioma cells increases gradually alongside the progression of tumor grade. Glioblastoma (Grade IV), which is the most malignant subtype, accounts for 56.6% of all gliomas (2). Despite the developments in treatment in recent years, the prognosis of glioblastoma patients has remained very poor, with a median life expectancy of less than 2 years (3-5). Therefore, it is crucial to exploit the underlying mechanism of tumorigenesis and develop effective therapeutic strategies for malignant glioma.
Glycosylation is one of the most common types of post-translational modification, and is critical for the biological functions of proteins (6,7). More than half of all known proteins are affected by glycosylation modification. Aberrant glycosylation is linked to several human diseases, including cancer. Functionally, glycosylation regulates many aspects of cancer cell biology, such as cellular signaling, tumor invasion, and metastasis (8-11). In brain cancer, especially in glioblastoma, salient alterations of glycosylation have been described (12-14). Inhibition of protein glycosylation provides a new strategy for glioblastoma treatment. A novel inhibitor of N-linked glycosylation has been demonstrated to inhibit the glycosylation and phosphorylation of multiple receptor tyrosine kinases (RTKs), which in turn retard tumor growth in the glioblastoma models (15,16).
Uridine diphosphate (UDP)-N-acetylglucosamine pyrophosphorylase-1 (UAP1) is an enzyme that catalyzes the synthesis of a sugar donor, UDP-N-acetylglucosamine (UDP-GlcNac), for glycosylation (17). UAP1-like-1 (UAP1L1) is a paralog of UAP1, and they share 59% identical sequences. Hill et al. have reported that UAP1L1 gene methylation is associated with relapse-free survival (RFS) in breast cancer (18). More recently, Lai et al. demonstrated that UAP1L1 is a critical factor for protein glycosylation and promotes proliferation in human hepatoma cells (19). However, there is very limited research on the functional role of UAP1L1. By analyzing the publicly available Gene Expression Omnibus (GEO) database, we found that UAP1L1 displayed a significant change between paired glioma and normal brain tissues (GSE41031, P=0.0035). We speculated that UAP1L1 might participate in the development of glioma.
In this study, for the first time, we investigated the expression and functional role of UAP1L1 in glioma. We found that UAP1L1 expression was obviously upregulated in the glioma tissues. The increased UAP1L1 expression level was associated with higher tumor grades and poorer patient prognoses. We then verified that UAP1L1 knockdown significantly inhibited proliferation of glioma cells both in vitro and in vivo. The results indicated that UAP1L1 could be a potential therapeutic target of malignant glioma.
We present the following article in accordance with the Animal Research: Reporting of In Vivo Experiments (ARRIVE) reporting checklist (available at http://dx.doi.org/10.21037/atm-20-2809).
Methods
Tissue microarrays
A tissue microarray that included 160 glioma samples was purchased from Outdo Biotech Co. Ltd. (chip no. XT18-004; Shanghai, China). A tissue microarray that included 23 normal brain samples was purchased from Alenabio Co., Ltd (chip no. GLN241; Xi’an, China). The array dot diameter was 1.5 mm, and each dot represented a tissue sample from 1 individual specimen of confirmed glioma. Their diagnoses were independently reviewed by 2 pathologists and classified by the 2016 WHO criteria. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the Ethics Committee of Xiangya Hospital of Central South University (2017121173). Informed consent was provided by all participants.
Cell lines and culture conditions
Human glioblastoma cell lines including U251 and U87 were originally obtained from the Chinese Academy of Sciences Cell Bank (Shanghai, China). These cells were routinely cultured in high glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS).
Animals
Four-week-old female BALB/c-nude mice of (Shanghai Ling-Chang Laboratory Animal Technology Co. Ltd., Shanghai, China) were housed in pathogen-free conditions and fed ad libitum. All animal experiments were conducted in accordance with the Chinese guidelines for the care and use of animals and approved by the Ethics Committee of Xiangya Hospital of Central South University (No. 2017121187).
Immunohistochemistry (IHC)
Tissue sections were cut and mounted on slides. IHC was performed as described previously (20). After deparaffination and rehydration, antigen retrieval was performed by immersing sections in 10 mM citrate buffer (pH 6.0) for 2 minutes at 100 °C. Endogenous peroxidase activity was blocked with 0.3% H2O2, and non-specific antigens were blocked with normal goat serum. The slides were incubated with primary antibody against human UAP1L1 (1:200 dilution, Abcam, Cambridge, MA, USA) overnight at 4 °C. After incubating with a secondary antibody, 3,3’-diaminobenzidine (DAB) chromogen was added to detect the IHC signal. Phosphate-buffered saline (PBS) was used as negative control to replace the primary antibody.
For statistical analysis, the expression level was recorded as immunoreactive score (IRS), which was calculated by multiplying the score of the staining intensity and the percentage of positive cells. The UAP1L1 staining intensity was scored as 0–3 (0, negative; 1, weak; 2, moderate; 3, strong). The percentage of UAP1L1-positive–stained cells was scored as follows: 1 (0–25%), 2 (26–50%), 3 (51–75%), and 4 (76–100%). According to the IRS, the UAP1L1 staining pattern was classified as having low (IRS 0–3) and high (IRS 4–12) expression.
Quantitative real-time polymerase chain reaction
Endogenous UAP1L1 expression in U251 and U87 glioma cells was detected by quantitative real-time polymerase chain reaction (qRT-PCR). Trizol reagent (Sigma-Aldrich, St. Louis, MO, USA) was used to extract RNA from the U251 and U87 cells. The complementary (cDNA) was synthesized from total RNA using an Expand Reverse Transcriptase Kit (Vazyme Biotech, Nanjing, China). The primer sequences for UAP1L1 were as follows: forward 5’-GGAGCGGAAAGACAAAGTTGC-3’ and reverse 5’-CACAGAAGCCGATGAAGACAGG-3’; glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a standard. The amplification was performed under the following conditions: 1 cycle of 95 °C for 30 seconds, 45 cycles of 95 °C for 5 seconds, and 60 °C for 30 seconds. The messenger RNA (mRNA) expression was normalized to the level of GAPDH mRNA and calculated by the 2−ΔΔCt method. Diethyl pyrocarbonate (DEPC) water was used as negative control in place of the template cDNA.
Lentiviral shRNA vector construction and infection
For knockdown of expression, UAP1L1-specific short hairpin RNA (shRNA) was synthesized and cloned into a lentiviral vector BR-V108 (Genechem, Shanghai, China). The constructed vector targeting UAP1L1 was named shUAP1L1, and the corresponding empty vector (BR-V108) was used as a negative control and named shCtrl. To avoid any off-target effects, 3 shRNAs were prepared to knockdown UAP1L1.
The lentiviral particles were produced by co-transfecting shUAP1L1/shCtrl with pHelper system (Genechem, Shanghai, China) into 293T cells. Lentivirus supernatants were harvested and used to infect U87 and U251 cells. Infection efficiency was observed under a fluorescence microscope after 72 hours. The knockdown of genes was confirmed by western blot.
Western blot analysis
The western blot analysis procedure was carried out as described previously (21). The total proteins were extracted from cell lines using radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime Biotechnology, Shanghai, China). Proteins were separated by 8% to 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA, USA). After blocking with 5% skim milk, the membranes were incubated with primary anti-UAP1L1 (1:1,000 dilution, Abcam, USA) and anti-GAPDH (1:2,000 dilution; Abcam, USA) overnight at 4 °C. Afterwards, the secondary antibody (1:2,000 dilution; Beyotime Biotech, Shanghai, China) was added and incubated for 2 hours at room temperature. Protein bands were detected using an ECL Plus detection kit (Amersham, GE Healthcare, USA).
Cell proliferation and colony formation assay
Cell proliferation was evaluated by using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Sigma-Aldrich). U251 and U87 cells infected with shUAP1L1 or shCtrl were seeded in 96-well plates at a density of 2,000 cells/well and cultured at different time intervals. The cells were then treated with MTT reagent for 4 hours at 37 °C, supernatants were removed, and dimethyl sulfoxide (DMSO; Sigma-Aldrich) was added to dissolve precipitates. Finally, the optical density (OD) was measured with a microplate reader (Tecan Group Ltd., Männedorf, Switzerland) at a wavelength of 490 nm.
For colony formation assays, cells infected with shUAP1L1 or shCtrl were seeded onto 6-well plates at a density of 600 cells/well and cultured for 14 days, and each experimental group was plated in 3 wells. After fixation with 4% paraformaldehyde for 30 minutes, the cell colonies were treated by Giemsa staining (Dingguo Biotech, Shanghai, China) for 15 minutes. A colony was defined as a formation consisting of more than 50 cells.
Cell apoptosis analyzed by flow cytometry
Apoptosis was assessed using an Annexin V-APC–labeled apoptosis detection kit (eBioscience, San Diego, CA, USA). Briefly, cells infected with shUAP1L1 or shCtrl were seeded in 6-well plates and cultured in DMEM with 10% FBS. When the cells were over 85% in confluence, they were harvested and stained with APC-labeled Annexin V in the dark for 30 minutes, and then these samples were analyzed by flow cytometry assay (FCA).
Human apoptosis antibody array
To determine the changes of apoptosis-related proteins response to the infection with shUAP1L1, a Human Apoptosis Antibody Array kit (Abcam, Cambridge, MA, USA) containing 43 spots was used. Briefly, the membranes were incubated with blocking buffer at room temperature for 30 minutes. Lysate of cells was then added and maintained at 4 °C overnight. After thorough washing, the membranes were incubated with biotin-conjugated antibodies for 2 hours followed by addition of streptavidin-horseradish peroxidase (HRP). The signals of membranes were detected using an ECL Plus kit.
In vivo xenograft model in nude mice
The xenograft nude mouse model was established as previously described (22). Briefly, 10 BALB/c‐nude mice were divided randomly into 2 groups: a control group (shCtrl) and a shRNA group (shUAP1L1). A total of 2×106 U87 cells infected with shUAP1L1 or shCtrl were injected subcutaneously into the upper right flank region of the nude mouse and permitted to grow until palpable. Then, tumors were measured twice per week with a vernier caliper, and tumor volumes were calculated with the following formula: volume = tumor length×width2×0.52. Tumor-bearing mice were examined with an in vivo imaging system (Berthold Technologies, Bad Wildbad, Germany) at day 28 after implantation; the mice were then sacrificed and tumors were weighed and photographed. The removed tumor tissues were used for Ki67 IHC.
Statistical analysis
Data are presented as mean ± SD and were analyzed using SPSS 17.0 software (IBM Corp., Armonk, NY, USA). All the cell experiments were performed independently at least 3 times, and statistical differences between groups were determined by Student’s t-test and Fisher’s exact test. The prognostic significance analysis was performed using the Kaplan-Meier method and log-rank tests. A P<0.05 was considered statistically significant.
Results
UAP1L1 expression was significantly upregulated in glioma tissues and correlated with poor patient survival
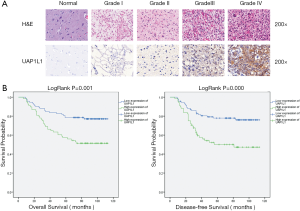
To determine the expression level of UAP1L1 in glioma, IHC staining was performed in tissue microarrays of 160 gliomas and 24 normal brain tissues. As shown in Figure 1A, UAP1L1 staining was primarily located in the cytoplasm. All normal brain tissues expressed UAP1L1 at a low level. A high level of UAP1L1 was observed in glioma samples, especially in high grade gliomas (grade III and grade IV). The expression of UAP1L1 was significantly increased in the glioma tissues when compared with the normal brain tissues (Table 1, P<0.001).

Full table
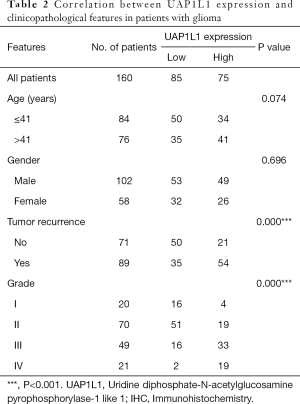
The correlation of UAP1L1 expression with clinicopathological parameters is summarized in Table 2. Mann-Whitney U analysis revealed that the expression of UAP1L1 in the glioma tissues was significantly associated with patient age, tumor grade, and recurrence. There was no significant association between UAP1L1 expression and gender. Spearman’s correlation analysis demonstrated a positive correlation of UAP1L1 expression with patient age, tumor grade, and recurrence.
Full table
Kaplan-Meier survival curves were constructed to further investigate whether increased UAP1L1 expression correlated with a worse prognosis in glioma patients (n=160, follow-up time 6–113 months). The data demonstrated that high UAP1L1 expression correlated with both worse overall and disease-free survival in glioma patients (Figure 1B, P=0.001, P<0.001 respectively).
Knockdown of UAP1L1 inhibited the proliferation and colony formation capacity of glioma cells in vitro
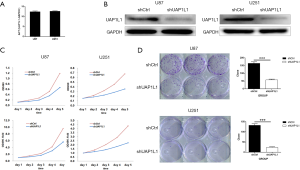
The endogenous expression of UAP1L1 in glioblastoma cells was detected using qRT-PCR. There was an abundant expression of UAP1L1 both in U251 and U87 cell lines (Figure 2A). Therefore, the lentiviral shRNA vector (shUAP1L1) was constructed and used to infect U251 and U87 cells to knockdown the expression of UAP1L1. In order to avoid any off-target effects, another 2 shRNAs targeting UAP1L1 were constructed (results not shown). Western blot analysis confirmed that UAP1L1 expression was obviously downregulated in U251 and U87 cells after infection with shUAP1L1 (Figure 2B). The MTT assay was applied to measure cell viability. The knockdown of UAP1L1 significantly decreased the growth rate of U251 and U87 cells (Figure 2C). We also explored the effects of UAP1L1 on cell proliferation using a colony formation assay. Knockdown of UAP1L1 significantly suppressed the colony formation capacity of glioblastoma cells when compared with the control (Figure 2D) (P<0.001). These findings suggested that UAP1L1 may contribute to the growth of glioblastoma cells in vitro.
Knockdown of UAP1L1 induced glioma cell apoptosis in vitro
Flow cytometry analysis was performed to examine the effect of UAP1L1 knockdown on cell apoptosis. The proportion of apoptotic cells in the shUAP1L1 infection group was markedly increased compared with the control (Figure 3A) (P<0.001), suggesting the knockdown of UAP1L1 promoted U87 and U251 cell apoptosis.
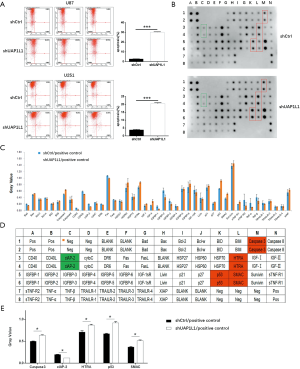
The Human Apoptosis Protein Array was used to further illuminate the expression of apoptosis-related proteins in response to UAP1L1 knockdown. As shown in Figure 3B,C,D,E, the caspase 3, p53, high temperature requirement A (HTRA) and second mitochondria-derived activator (SMAC) levels were obviously increased in the shUAP1L1 infection group, while cellular inhibitor of apoptosis protein-2 (cIAP-2) was decreased, suggesting that knockdown of UAP1L1 could induce apoptosis signaling in glioblastoma cells through the upregulation of caspase 3, HTRA, p53, and SMAC expression, and the downregulation of clAP-2 expression.
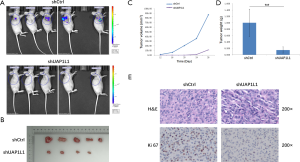
Knockdown of UAP1L1 inhibited glioma cell growth in vivo
A nude mouse glioma xenograft model was established to evaluate the effect of UAP1L1 knockdown on the growth of glioblastoma cells in vivo. The UAP1L1-depleted U87 cells (shUAP1L1 group), and the control cells (shCtrl group) were successfully transplanted into the back of the nude mice. After 28 days, the bioluminescence signal intensity in the shUAP1L1 group was obviously lower than that of the control (Figure 4A). The mean volume and weight of subcutaneous tumors in the shUAP1L1 group were obviously smaller and lighter than those in the control group (Figure 4B,C,D, P<0.01). Next, we investigated the expression of Ki67, which is a marker of proliferation, in the xenograft tumor tissues. The percentage of Ki67-positive cells was markedly decreased in the UAP1L1 knockdown group (Figure 4E). Taken together, these results demonstrated that UAP1L1 knockdown attenuated glioblastoma cell proliferation in vivo.
Discussion
The main purpose of this study was to investigate the biological function of UAP1L1 in glioma. Using IHC analysis, we found that UAP1L1 was notably upregulated in glioma tissues compared with normal brain tissues. The increased expression of UAP1L1 was correlated with higher glioma grade and poorer survival in glioma patients. Due to the high level of UAP1L1 in U87 and U251 glioblastoma cell lines, we constructed a lentivirus-mediated shRNA to knock down the expression of UAP1L1 in U87 and U251 cells. Our results showed that downregulation of UAP1L1 could suppress the proliferation and cell colony formation capacity of U87 and U251 cells. The depletion of UAP1L1 could also promote cell apoptosis by downregulating apoptosis-related proteins, including caspase3, HTRA, p53, and SMAC, and downregulating clAP-2. In addition, the xenograft model further confirmed that knockdown of UAP1L1 significantly inhibited the growth of glioma. Therefore, UAP1L1 may play an oncogene-like role in the tumorigenesis of glioma.
The specific mechanism through which UAP1L1 regulates proliferation and apoptosis of glioma cells remains unclear. UAP1L1 is a paralog of UAP1, while UAP1 is an enzyme that directly participates in the synthesis of sugar donor for glycosylation (17). It has been reported that UAP1 was overexpressed in prostate cancer and contributed to the growth and survival of cancerous cells (23), while upregulating the expression of UAP1 was found to promote Kirsten rat sarcoma viral oncogene homolog (KRAS)-driven lung tumorigenesis by elevating protein O-GlcNAcylation modification (24). Novel anticancer agents have been discovered to inhibit the key enzymes, including UAP1, in glycosylation (25). Despite having established a 59% sequence identity for UAP1, we know very little about the function of UAP1L1. Lai et al. demonstrated that UAP1L1 was significantly upregulated in hepatocellular carcinoma tissues and that a high level of UAP1L1 expression predicted a poor prognosis. The authors stated that “UAP1L1 promotes human hepatoma cell growth both in vitro and in vivo”. They also demonstrated that UAP1L1 was critically involved in protein glycosylation, but it functioned distinctly from UAP1 (19). Aberrant glycosylation was closely related to the development and progression of several cancers, including that of the brain (26,27). Some highly glycosylated proteins, such as MUC4 and ST3GAL1, have been found to be overexpressed in glioblastoma and to play roles in tumorigenesis and invasion (28,29). We thus propose that UAP1L1 may promote glioma cell proliferation through regulating the glycosylation status of some key proteins. However, glycosylation alterations in cancers are highly complex, and can include the aberrant expression of enzymes in glycan biosynthesis and post-synthetic modification (26). Clarifying the specific molecular mechanism of UAP1L1 through further experimentation may yield valuable insights.
Conclusions
Our findings demonstrate UAP1L1 plays an oncogene-like role in glioma, especially in high grade glioma. Therefore, UAP1L1 may be a potential biomarker for diagnosis and prognosis, and be a promising therapeutic target for gliomas with high levels of UAP1L1.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (No. 81672759).
Footnote
Reporting Checklist: The authors have completed the Animal Research: Reporting of In Vivo Experiments (ARRIVE) reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-2809
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-2809
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-2809
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-2809). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The human study was approved by the ethics committee of the Xiangya Hospital of Central South University (No. 2017121173) and informed consent was provided by all participants. Animal experiments were performed under a project license (No. 2017121187) granted by the ethics committee of the Xiangya Hospital of Central South University, in compliance with Chinese guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 2016;131:803-20. [Crossref] [PubMed]
- Ostrom QT, Gittleman H, Truitt G, et al. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2011-2015. Neuro Oncol 2018;20:iv1-86. [Crossref] [PubMed]
- Stupp R, Taillibert S, Kanner AA, et al. Maintenance Therapy With Tumor-Treating Fields Plus Temozolomide vs Temozolomide Alone for Glioblastoma: A Randomized Clinical Trial. JAMA 2015;314:2535-43. [Crossref] [PubMed]
- Weller M, van den Bent M, Tonn JC, et al. European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol 2017;18:e315-29. [Crossref] [PubMed]
- Zhang H, Wang R, Yu Y, et al. Glioblastoma Treatment Modalities besides Surgery. J Cancer 2019;10:4793-806. [Crossref] [PubMed]
- Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta 1999;1473:4-8. [Crossref] [PubMed]
- Moremen KW, Tiemeyer M, Nairn AV. Vertebrate protein glycosylation: diversity, synthesis and function. Nat Rev Mol Cell Biol 2012;13:448-62. [Crossref] [PubMed]
- Reis CA, Osorio H, Silva L, et al. Alterations in glycosylation as biomarkers for cancer detection. J Clin Pathol 2010;63:322-9. [Crossref] [PubMed]
- Drake RR. Glycosylation and cancer: moving glycomics to the forefront. Adv Cancer Res 2015;126:1-10. [Crossref] [PubMed]
- Chandler KB, Costello CE, Rahimi N. Glycosylation in the Tumor Microenvironment: Implications for Tumor Angiogenesis and Metastasis. CELLS-BASEL 2019;8.
- Gupta R, Leon F, Rauth S, et al. A Systematic Review on the Implications of O-linked Glycan Branching and Truncating Enzymes on Cancer Progression and Metastasis. Cells 2020;9:446. [Crossref] [PubMed]
- Veillon L, Fakih C, Abou-El-Hassan H, et al. Glycosylation Changes in Brain Cancer. Acs Chem Neurosci 2018;9:51-72. [Crossref] [PubMed]
- Wade A, Robinson AE, Engler JR, et al. Proteoglycans and their roles in brain cancer. FEBS J 2013;280:2399-417. [Crossref] [PubMed]
- Tuccillo FM, de Laurentiis A, Palmieri C, et al. Aberrant glycosylation as biomarker for cancer: focus on CD43. Biomed Res Int 2014;2014:742831 [Crossref] [PubMed]
- Baro M, Lopez SC, Quijano A, et al. Oligosaccharyltransferase Inhibition Reduces Receptor Tyrosine Kinase Activation and Enhances Glioma Radiosensitivity. Clin Cancer Res 2019;25:784-95. [Crossref] [PubMed]
- Wahl DR, Lawrence TS. No Sugar Added: A New Strategy to Inhibit Glioblastoma Receptor Tyrosine Kinases. Clin Cancer Res 2019;25:455-6. [Crossref] [PubMed]
- Peneff C, Ferrari P, Charrier V, et al. Crystal structures of two human pyrophosphorylase isoforms in complexes with UDPGlc(Gal)NAc: role of the alternatively spliced insert in the enzyme oligomeric assembly and active site architecture. EMBO J 2001;20:6191-202. [Crossref] [PubMed]
- Hill VK, Ricketts C, Bieche I, et al. Genome-wide DNA methylation profiling of CpG islands in breast cancer identifies novel genes associated with tumorigenicity. Cancer Res 2011;71:2988-99. [Crossref] [PubMed]
- Lai CY, Liu H, Tin KX, et al. Identification of UAP1L1 as a critical factor for protein O-GlcNAcylation and cell proliferation in human hepatoma cells. Oncogene 2019;38:317-31. [Crossref] [PubMed]
- Yang Z, Wang Y, Fang J, et al. Expression and aberrant promoter methylation of Wnt inhibitory factor-1 in human astrocytomas. J Exp Clin Cancer Res 2010;29:26. [Crossref] [PubMed]
- Liu Y, Wu C, Wang Y, et al. Loss of plexin-B3 in hepatocellular carcinoma. Exp Ther Med 2015;9:1247-52. [Crossref] [PubMed]
- Ou C, Sun Z, He X, et al. Targeting YAP1/LINC00152/FSCN1 Signaling Axis Prevents the Progression of Colorectal Cancer. Adv Sci (Weinh) 2019;7:1901380 [Crossref] [PubMed]
- Itkonen HM, Engedal N, Babaie E, et al. UAP1 is overexpressed in prostate cancer and is protective against inhibitors of N-linked glycosylation. Oncogene 2015;34:3744-50. [Crossref] [PubMed]
- Taparra K, Wang H, Malek R, et al. O-GlcNAcylation is required for mutant KRAS-induced lung tumorigenesis. J Clin Invest 2018;128:4924-37. [Crossref] [PubMed]
- Yang Y, Vankayalapati H, Tang M, et al. Discovery of Novel Inhibitors Targeting Multi-UDP-hexose Pyrophosphorylases as Anticancer Agents. Molecules 2020;25:645. [Crossref] [PubMed]
- Lemjabbar-Alaoui H, McKinney A, Yang YW, et al. Glycosylation alterations in lung and brain cancer. Adv Cancer Res 2015;126:305-44. [Crossref] [PubMed]
- Tang L, Chen X, Zhang X, et al. N-Glycosylation in progression of skin cancer. Med Oncol 2019;36:50. [Crossref] [PubMed]
- Li W, Wu C, Yao Y, et al. MUC4 modulates human glioblastoma cell proliferation and invasion by upregulating EGFR expression. Neurosci Lett 2014;566:82-7. [Crossref] [PubMed]
- Chong YK, Sandanaraj E, Koh LW, et al. ST3GAL1-Associated Transcriptomic Program in Glioblastoma Tumor Growth, Invasion, and Prognosis. J Natl Cancer Inst 2015;108:djv326 [PubMed]
(English Language Editor: J. Jones)


