
MicroRNA and diabetic retinopathy—biomarkers and novel therapeutics
Introduction
Diabetic retinopathy (DR) accounts for ~80% of legal blindness in persons aged 20–74 years and causes enormous social and health burdens (1). In 2010, ~7.7 million persons in the US had DR (2,3), which is expected to rise to ~16 million by 2050 (2). The pathogenesis of DR is complex but mainly results from adverse metabolic effects of chronic hyperglycemia, which cause retinal microvascular damage and lead to retinal damage and ischemia. This ischemic environment stimulates the retina to produce growth factors, such as vascular endothelial growth factor (VEGF), which promote angiogenesis, vascular leakage, and inflammation, ultimately contributing to vision loss (4). Current therapies for DR include laser therapy, intravitreal injections of anti-VEGF agents, anti-inflammatory and corticosteroid therapy, and intraocular surgery. However, they are invasive, non-curative and alleviate vision impairment only temporarily. Thus, repeated treatments are often required.
The clinical challenge of treating DR
DR and associated ocular complications, such as macular edema and retinal neovascularization, are leading causes of vision impairment and blindness in the US and worldwide. Intravitreal injection of anti-VEGF agents can treat both diabetic macular edema and retinal neovascularization. The problems with current intravitreal anti-VEGF therapy include: (I) need for repeated treatments; (II) potential off-target effects on other, non-endothelial, cells; and (III) involvement of factors other than VEGF in pathogenesis of DR, which may limit or attenuate patient response to anti-VEGF agents. About 15–25% of DR patients do not respond to anti-VEGF therapy (5-7). Therefore, there is a great unmet need to improve treatments for DR.
A new approach to treating DR that can be developed is via precision medicine that targets novel epigenetic changes associated with DR. It contrasts with a one-size-fits-all approach currently used, in which disease treatment and prevention strategies are developed for the average person, with less consideration for the genetic differences among individuals. The need for such new therapeutic approaches is clear and compelling. Targeting specific microRNAs (miRNAs) is one such approach to manage diabetic retinopathy.
The unique potential of targeting specific miRNAs in DR
This review explores the potential utility of miRNAs, small non-coding forms of RNA implicated in human diseases, in diagnosis and management of DR (8,9). MiRNAs post-transcriptionally regulate gene expression by degrading or blocking the translation of messenger RNA (mRNA) targets. MiRNAs generally silence the expression of target genes by binding to specific sites on the genes 3’ untranslated region (UTR) and recruiting a silencing complex that blocks translation (10). Besides their presence in tissues, miRNAs circulate in the bloodstream in a highly stable, extracellular form and can serve as easily accessible blood-based biomarkers (11). We will discuss the roles of individual specific miRNAs and their target molecular pathways in DR and assess their potential applications for treating DR. A novel therapeutic approach that can be explored is modulating pathways critical to pathogenesis DR using miRNAs or anti-miRNAs (anti-miRs), chemically-engineered to block miRNA molecules from binding to their specific target sites on mRNA. Alternatively, endogenous miRNA actions may be silenced by targeting binding sites on mRNAs, as well as long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs), which are natural regulators of the regulators.
MiRNAs are long-distance communicators and reprogramming agents
Several miRNA species act as master regulators of development, inflammation, and aging (12-15). Such miRNAs secreted extracellularly into bodily fluids, migrate in small membranous extracellular vesicles (EVs) or exosomes, and are then endocytosed to act as long-distance communicators and reprogramming agents on distant cells. MiRNAs represent a completely novel paradigm in cellular and tissue signaling and interaction, and understanding their endogenous roles and potential applications concerning DR may lead to novel approaches to diagnosis and treatment.
Altered intracellular expression levels of miRNAs play a role in DR
There are many excellent reviews about miRNAs (16-18) describing their biogenesis, processing, and regulation of gene expression, so we will not go into those details. The majority of the miRNAs are present intracellularly, while a special class of miRNAs have been identified in bodily fluids and named circulatory miRNAs or extracellular miRNA. They were initially identified in blood and then subsequently in all bodily fluids, including ocular aqueous and vitreous humor. Their dysregulation is present in many diseases and disorders, and they have been identified as early biomarkers. Several examples of identified intracellular DR biomarker miRNAs are presented in Table 1. Overexpression of miR-21 has been seen to play a pivotal role in the pathogenesis of DR by contributing to diabetes mellitus (DM)-induced endothelial dysfunction as well as low-grade inflammation (19). In a DR rat retina, overexpression of miR-216a protects against human retinal microvascular endothelial cell (HRMEC) injury in DR by suppressing the NOS2/JAK/STAT axis (20). At the same time, miRNA-29b-3p promotes HRMEC apoptosis via blocking SIRT1 in DR (21). The upregulation of miR-203a-3p might inhibit retinal neovascularization in oxygen-induced retinopathy (OIR) rat model via targeting VEGFA and HIF-1α (22). MiR-183 silencing inhibits cell growth and tube formation in vascular endothelial cells of DR rats via the PI3K/Akt/VEGF signaling pathway by upregulating BTG1 (25). The relationship between miR-200b-3p and MALAT1 or vascular endothelial growth factor A (VEGFA) was validated in a DR mouse model that was induced by streptozotocin injection and a high-glucose/high-fat diet and in retinal microvascular endothelial cells (RMECs) (24).
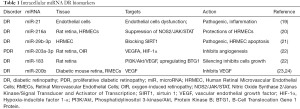
Full table
Single nucleotide polymorphism of miRNA and target genes
Genetic variations, mostly single-nucleotide polymorphisms (SNPs), are changes in a single nucleotide in a sequence of miRNA (26). Variations in miRNA transcripts can cause structural mismatches in hairpin loops, essential and conserved structural features for miRNAs, resulting in miRNA expression changes, abnormal function, and dysregulation of target genes. SNPs in the miRNAs seed recognition region can cause changes in stability of miRNA:mRNA binding sites or generate new binding sites, which can change the target specificity of miRNA (27) (Figure 1). That kind of change in gene regulation can have far-reaching consequences for cellular physiology, and there are a few miRNA SNPs identified that show association with DR (Table 2).
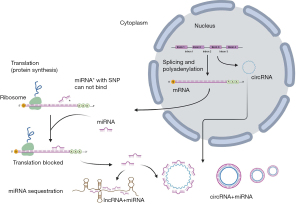

Full table
SNP in the MIR146a gene (rs2910164) is significantly associated with microvascular protection in DR patients with type 2 diabetes mellitus (T2DM) (28). Common SNP polymorphisms in miR-146a and miR-155 variations provide a protective effect for patients with type 1 diabetes mellitus (T1DM) by negatively regulating inflammation. In miR-126, the A allele of rs4636297, known to be the non-functional allele for post-translational regulation of the miRNA, is associated with severe non-proliferative diabetic retinopathy (NPDR) or proliferative diabetic retinopathy (PDR) (29).
A common polymorphism in MIR449b is significantly associated with a decreased risk of PDR and sight-threatening DR in Caucasian patients with T1DM (30). MiRNA-449b is part of a family of miRNAs involved in cell proliferation, inflammation, and angiogenesis and are important hypoxia-regulated miRNAs. MiRNA-449a and 449b have been studied in retinal tissues of mice (32,33). In an oxygen-induced mouse model of retinal ischemia, miRNA 449a was significantly downregulated, which is consistent with a potential protective role against retinal ischemia (33).
Circulating miRNA-155 (miR-155) is associated with T2DM, and the rs767649 polymorphism in the pre-MIR155 gene is associated with DR. MiR-155 is a regulator of immune system cell proliferation, cell differentiation, and effector responses, such as cytokine and antibody production. MiR-155 was shown to be a positive regulator of insulin sensitivity and required for normal blood glucose homeostasis. MiR-155 is also expressed in normal murine retina and retinal endothelial cells (31).
Circulatory miRNAs potential as early diagnostic tools for DR
Circulatory miRNAs have been shown to be differentially expressed in people with DM in serum and plasma studies, urine, and many other bodily fluids (34), suggesting that miRNAs could also serve as novel biomarkers in detecting or predicting the overall progress of DM and perhaps the progress of retinopathy from mild to sight-threatening (35-37). Although, in order to identify robust biomarkers, it is essential to establish miRNA profiles of ocular fluids and plasma and correlations of dysregulations of miRNAs in aqueous, vitreous, and plasma.
Analysis of circulating miRNAs from serum or plasma samples of DM patients, both with and without DR, showed altered expression levels of many miRNAs throughout different patient populations (38) (Table 3). The serum expression of miR-210 was upregulated in DR patients compared with DM patients with no retinopathy and healthy controls. Furthermore, proliferative DR (PDR) patients had higher levels than non-proliferative DR (NPDR) patients. The differential upregulation suggests that increased serum miR-210 could be used to identify patients with higher risk for PDR from NPDR patients (39). Two miRNAs mostly associated with the risk of DR in patients with T1DM are miR-27b and miR-320a (40). An anti-angiogenic miRNA, miR-221, has been identified in serum as a biomarker for DR in patients with T2DM and PDR, and it was involved in the DM physiopathology and macrovascular complications associated with T2DM (46-49). It was increased in serum, together with Ang II and VEGF (46). Potential biomarkers let-7a and miR-151 in serum have been confirmed by RNA seq for late-stage and early-stage DR in patients with T2DM (50). Besides, other circulating miRNAs, including miR-126, miR-150, miR-155, and miR-200b, were also found dysregulated in DR patients, as well as in preclinical animal models of DR [(50), and references therein]. These findings indicate the complex regulation of miRNAs in DR and the great potential of miRNAs as biomarkers of DR.
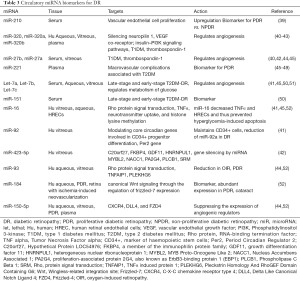
Full table
The studies of miRNAs in vitreous of human DR patients found biomarkers such as let-7c, miR-16, miR-92a, and miR-320a, b [(41), and refs therein]. Some of the miRNA candidates identified in other studies are from the same families (let-7, miR-320), although not exactly the same member. Several miRNAs (including miR-27a, miR-93, and miR-150) were identified in vitreous as diagnostic and prognostic miRNAs for severe complications in patients with PDR (44).
Eight miRNAs, including miR-184, miR-150, miR-16, and miR-93, were identified in the aqueous of the patients as the most differentially expressed between PDR and controls (P>0.85), using next-generation sequencing (NGS). This differential expression of miRNA was predicted to regulate Rho protein signal transduction, neurotransmitter uptake, and histone lysine methylation (52).
Our comparison of circulatory miRNAs revealed that among 847 human miRNA probes on the Affymetrix miRNA microarrays, common miRNAs were present both in the aqueous and vitreous humor, as were a large number of unique miRNA, dependent upon DM type and DR severity. We identified several miRNAs that showed altered levels in the vitreous humor of human DR patients vs. that of healthy controls (52-54). In contrast, altered miRNA levels in aqueous humor did not appear to be a useful marker of miRNA abundance in vitreous humor or plasma. Still, a few potential candidates for common biomarkers stood out (45). We found a strong upregulation of let-7b in ocular fluids in T1DM-PDR and T2DM-PDR vs. controls. Other let-7 family members (let-7c, let-7d) were also upregulated in PDR, but more modestly. The let-7 family members also showed apparent dysregulation in more than one category of DR. Our second candidate miR-320b, was upregulated in vitreous of both T1DM-PDR 9-fold and in T2DM-PDR 8-fold. Other miR-320 family members, miR-320a, and miR-320c showed the same trend (45).
It is very encouraging to see that putative miRNA biomarker findings (45) have been confirmed by different groups using different techniques. However, some studies identify entirely different sets of biomarkers, as it happened with RNA-seq study of non-proliferative DR biomarkers for T2DM-DR patients in Chinese Han ancestry in serum (55). This result poses questions of the ethnic group variability and redundancy of the miRNA family member function. Therefore, more research is needed, and perhaps a variety of ethnic groups should be studied to address those questions.
Roles of lncRNAs in DR
LncRNAs are RNA transcripts of more than 200 nucleotides that do not encode proteins. LncRNA and miRNAs are subclasses of non-protein coding RNA transcripts. LncRNAs are key molecules in regulating several biological processes and complex diseases such as cancer and neurological disorders (56,57). LncRNAs function in several modes of action—in cis by regulating nearby genes, or in trans by modulating distantly located genes (58). Additionally, a mechanism that lncRNAs may regulate target gene expression via miRNA response element (MRE), known as competing endogenous RNA (ceRNA) has been described (59,60). They are also called “sponge” RNAs if they contain several complementary binding sites to a miRNA of interest, and, as with most miRNA target genes, a sponge’s binding sites are specific to the miRNA seed region, which allows them to block a whole family of related miRNAs (61) (Figure 1).
The dysregulation of lncRNAs in early DR was first characterized in 2014. Pathway analysis indicated activation of the axon guidance signaling pathway (62). Studies of individual lncRNAs playing a role in DR pathogenesis and progression followed, including Maternally Expressed 3 (MEG3) (63), Myocardial Infarction Associated Transcript (MIAT) (64,65), HOXA Distal Transcript Antisense RNA (HOTTIP) (66), Metastasis Associated Lung Adenocarcinoma Transcript 1 (MALAT1) (67,68), SOX2 Overlapping Transcript (Sox2-OT) (69), antisense non-coding RNA in the INK4 locus (ANRIL), a regulator of VEGF (70), and BDNF Antisense RNA (BDNF-AS) (71). There is also recently identified a regulatory network of 3 novel lncRNAs (MSTRG.15047.3, FRMD6-AS2, and AC008403.3) transthyretin attenuating HREC dysfunction in DR. In DR patients, MSTRG.15047.3 and AC008403.3 showed significantly relative higher expression in both aqueous humor and serum samples, compared with healthy controls, and FRMD6-AS2 was significantly down-regulated (72).
miRNA/lncRNA regulatory loops and target pathways in DR
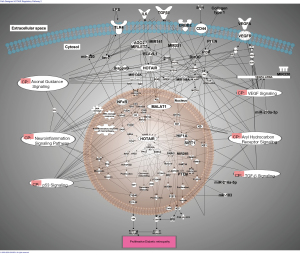
The regulatory loops of miRNA/lncRNAs—therapeutic strategies can be borrowed from natural regulatory loops to plan targeted therapy. Here are several examples of this type of regulatory network identified in diabetes and relevant to DR (Table 4, Figure 2).

Full table
H19 and miR-200b
The pathophysiology of DR is linked to high glucose levels and its effect on retinal microvascular tissues. The resulting endothelial injury changes the endothelial cell phenotype to acquire mesenchymal properties [i.e., endothelial-mesenchymal transition (EndMT)]. LncRNA H19 prevents EndMT in DR. LncRNA H19 is downregulated under high glucose conditions and regulates the expression of miR-200b. H19 and miR-200b regulate TGF-β1 signaling. Overexpression of H19 decreases TGF-β1 levels and affects the TGF-β1–MAPK–ERK1/2 signaling pathway [a Smad-independent pathway (SHC-transforming protein, GRB2, SOS)] by preventing pERK1/2 protein expression. In turn, this leads to an increase in endothelial markers and a reduction in mesenchymal markers, leading to the prevention of EndMT (73).
MIAT and miR-150
MIAT knockdown enhanced diabetes-induced retinal microvascular dysfunction and inhibited endothelial cell proliferation, migration, and tube formation in vitro. Further studies revealed that MIAT functioned as a competing endogenous (ce)RNA and formed a feedback loop with VEGF mRNA and miR-150-5p to regulate endothelial cell function (65).
MIAT and miR-29b
The high glucose promoted the binding activity between NF-κB and MIAT, while miR-29b controlled MIAT to regulate its expression. MIAT overexpression suppressed miR-29b, but promoted Sp1. The mechanism of cell apoptosis in DR might be associated with the regulation of MIAT; however, miR-29b acted as a biomarker regulated by MIAT and further regulated cell apoptosis in DR (64).
MALAT1 and miR-203a-3p
LncRNA MALAT1 acts as a transcriptional regulator for numerous genes, including some genes involved in cancer metastasis, cell migration, and in cell cycle regulation (provided by RefSeq, Mar 2015, IPA). LncRNA MALAT1 participates in DR pathological angiogenesis in OIR mouse model by sponging miR-203a-3p (76). LncRNA MALAT1 accelerates wound healing of diabetic mice transfused with modified autologous blood via the HIF-1α signaling pathway (77,78). MALAT1 regulates VEGFA, and it is regulated by several miRNAs, such as miR-133a, miR-133, miR-126a-5p (and other miRNAs w/seed AUUAUUA).
MALAT1 and miR-320
Basal endothelial sprouting is an indicator of angiogenesis and proliferation. It has been reported that MALAT1 regulates angiogenesis through modulating genes associated with the cell cycle in endothelial cells. Moreover, in human umbilical vein endothelial cells (HUVECs), MALAT1 has been shown to competitively bind to miR-320a, partly block the direct interaction between miR-320a and FOXM1, and lead to endothelial cell proliferation and angiogenesis, suggesting that lncRNA MALAT1 may function as a competing endogenous RNA (ceRNA) to promote angiogenesis (78).
HOTAIR and let-7
LncRNA HOTAIR promotes tumor growth, invasion and migration and epithelial to mesenchymal transition. MALAT1 and HOTAIR are key epigenetic regulators in DR (67,75,79). HOTAIR interacts with let-7, miR-320,VEGF signaling, and many other key pathways (Axonal Guidance Signaling, Aryl Hydrocarbon Receptor Signaling).
miRNAs therapeutic can target whole pathways involved in DR
Circulating miRNAs that are ubiquitously present in aqueous and vitreous ocular chambers target primarily genes regulated by VEGF, p53, and TGF-β (45). Some of the putative pathway-targeting miRNA identified in our studies are let-7 and miR-320.
Let-7
The analysis revealed that a partial interactome of the let-7 family of miRNAs and some of the target genes and biological pathways that they regulate are TGF-β, Insulin Receptor, Ap, and VEGF Receptor Signaling. These genes are critical regulators of oxidative stress, angiogenesis, inflammation, and apoptosis, highly relevant to DR (45).
miR-320
The miR-320 family is part of the cellular response to glucose stimulus and plays a role in apoptosis, migration, cell death, proliferation, and signaling in several diseases, including non-insulin-dependent diabetes mellitus (IPA and refs therein). According to the IPA summary, the miR-320 family regulates multitude of genes, including IGF1 and TGF-β, and it is regulated by p53 and Smad2/3, which is downstream from TGF-β. Further IPA analysis of pathways targeted by both let-7 and miR-320 family have identified pathways connecting VEGF and TGF-β (Figure 2).
Pathway analysis
Our emerging results from the IPA pathway analysis of the most dysregulated miRNAs in the vitreous suggested that their regulatory targets are VEGF and TGF-β pathways (Figure 2). Current DR therapies have targeted VEGF for inhibition using antibodies, but the concept of targeting pathways, especially the regulatory TGF-β pathway, is novel and advantageous. TGF-β and its signaling pathway play important roles in angiogenesis, endothelial cell proliferation, adhesion, and extracellular matrix deposition (80,81), and are implicated in DR pathogenesis by disrupting angiogenesis and the blood-retinal barrier (82).
MiRNA as novel therapeutics
MiRNAs represent a novel and attractive target and possible therapeutic agent which could manipulate pathological pathways in the eye. MiRNAs targeting whole pathways of human disease provide a new and potentially powerful approach for therapeutic intervention against DR. There is a need to exploit the potential utility and promise of miRNAs, small non-coding forms of RNA implicated in human diseases (8,9). The expression of miRNAs is altered in various diseases, and it is now feasible to manipulate miRNA expression by delivering miRNAs similar to the use of antisense mRNAs and RNAi (widely used techniques for investigating gene function and in gene therapy (83). As the activation of oncogenes could cause cancer, artificial antisense miRNAs could be synthesized and employed to block their targeted oncomirs to prevent the formation of cancer. However, various critical prerequisite data must be available, for example, identification of tissue signature miRNAs, their mechanism of action, applicability by RNAi, delivery of miRNAs, and their active form in vivo. Once this information is available, miRNA will have a bright future and become a popular therapeutic tool. The process of building miRNA therapeutics is similar to drug discovery and development, and its detailed steps are covered in great detail in other reviews (84,85).
There are two main strategies to manipulate miRNAs, which are dependent on whether the targeted miRNA expression needs to be downregulated or to re-introduce to restore loss of function. Here we will highlight two approaches that we find very promising for delivering miRNA therapeutics.
Nuclease-protected miRs and anti-miRs
Naturally-occurring DNA or RNA are limited as exogenous agents owing to properties such as poor binding affinity and degradation by nucleases. Chemically-modified nucleic acids can overcome some such limitations. Among these, locked nucleic acid (LNA) modification has proven uniquely useful. Oligos that contain LNAs show enhanced stability and nuclease resistance, very high binding affinities, and excellent specificity toward complementary RNA or DNA oligonucleotides. These have been used in multiple nucleic acid-based therapeutic strategies both in vitro and in vivo (86,87). In vivo, LNA anti-miR can inhibit target molecules up to 15 weeks in live animals (88-91). They tested the potency and duration of RNA silencing by LNA GapmeRs in different tissues by systemically administering Antisense LNA GapmeRs targeting the highly and ubiquitously expressed Malat1 lncRNA in mice. Samples of 12 different tissues up to 15 weeks after the last dose revealed efficient and durable Malat1 knockdown (>5 wk after the last dose) in a broad range of tissues (89), suggesting suitability for use targeting our novel DR marker candidates.
Delivery of miRNAs to the target tissue
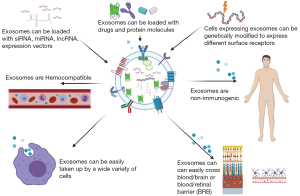
The high relative stability of miRNA in common clinical tissues and biofluids (e.g., plasma, serum, vitreous, etc.) results from their release from cells in the protective encapsulated form within microvesicles or exosomes and bound to proteins such as argonaute 2 or packaged within lipid particles (92). Such exosomes represent a promising delivery vehicle for novel drugs and gene therapy (93) (Figure 3). These lipid vesicles are secreted by all cells and can deliver proteins and RNA (94). The field of research what represents true microvesicles, EVs or exosomes is very active area of study. EVs are cell-derived membrane particles ranging from 30 to 5,000 nm in size, including exosomes, microvesicles, and apoptotic bodies (95,96). In this review, we are referring to the smallest of these EVs, microvesicles/exosomes, and we will call them exosomes, for the brevity’s sake. There is a strong interest for exosomes to be engineered for effective miRNA delivery, based on recent findings that exosomes derived from dendritic cells transmitted target-specific siRNA to the brain, a paradigm-shifting advance for prospective use of nanosized exosomes as a delivery system (97). In vivo targeting methods via other nanovesicles such as liposomes exist, but this novel exosome-based delivery approach is safer, as it doesn’t involve cells, and exosomes carry very low immunogenicity. Exosome-based delivery of therapeutics has the potential to conquer major hurdles, including cargos delivery across impermeable biological barriers (e.g., the blood-brain, blood-retinal, and blood-ocular barriers), biocompatibility, metabolic stability, target specific delivery, and has enhanced the efficacy of loaded therapeutic agents (97).
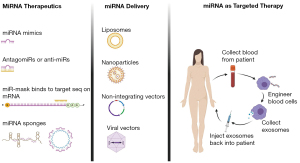
Other delivering methods for miRNAs
Several other methods for miRNA delivery are used in practice, such as AAV vectors, plasmid, piggybacks expression vectors, nanoparticles such as CC9 with a tumor-targeting and penetrating bifunctional peptide, peptide nucleic acid- artificial peptide polymer that binds to target nucleotide sequence are best covered in review articles (84). Different therapeutic miRNA strategies, delivery vehicles, and one of the therapeutic strategies for targeted therapy are summarized in Figure 4.
Challenges
However, although a plethora of miRNA-based compounds has been investigated in preclinical studies, only a minority of these have moved to clinical development. Challenges concerning a proper target selection, stability in body fluids, and specificity of target binding, as well as off-target effects, remain to be addressed in the future to optimize the in vivo delivery and efficiency of miRNA-based therapeutics. As miRNAs are implicated in virtually all physiological and pathological processes, a huge therapeutic potential is expected from miRNA-based constructs.
miRNAs as potential applications to yield novel therapies for DR—main points
- Novel therapeutic option for DR non-responders to VEGF therapy. About 15–25% of DR patients have a suboptimal response to anti-VEGF treatment. This approach would offer therapeutic alternatives to this significant group of patients who are resistant to anti-VEGF therapy.
- Lowering the cost of therapeutics. Small nucleic acids, miRNAs, and anti-miRNAs, are easy to synthesize. This ease of synthesis is an advantage over antibody therapy, which is the base of current anti-VEGF treatment. This synthesis advantage could result in a significantly lowered price of therapeutic production.
- Targeting multiple genes with one miRNA—pathway targeting. One miRNA or anti-miRNA targets multiple genes, frequently in the same pathway, and the signal amplifies immensely. Therefore, with one small nucleic acid molecule, it might become possible to modulate the whole signaling pathway.
- Combination therapy of multiple miRNAs and anti-miRNAs—a novel approach via combination therapy, targeting multiple pathways with multiple drugs, may achieve synergistic angiostatic activity that can circumvent DR pathologic angiogenesis problems.
- Exosome-assisted miRNA therapy would impart all the benefits of stem cell therapy, without the risks of transferring live cells into the eye. Ultimately, it is expected that new approaches mentioned in this review may create a stepping stone towards non-invasively delivering therapeutics, as exosome-encapsulated miRNAs by intravenous injections or via eyedrops.
Acknowledgments
Funding: This research was supported by the National Institutes of Health Grant from National Eye Institute [5R01EY021537 to ZSM], Innovative Development Grant (IDEA) from UC Davis to ZSM, an unrestricted grant from Research to Prevent Blindness (RPB) to UCD Dept. of Ophthalmology, and Barr Family Foundation grant (to LSM at Department of Ophthalmology at UC Davis).
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Susanna S. Park) for the series “Novel Tools and Therapies for Ocular Regeneration” published in Annals of Translational Medicine. The article has undergone external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-20-5189). The series “Novel Tools and Therapies for Ocular Regeneration” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bourne RR, Stevens GA, White RA, et al. Causes of vision loss worldwide, 1990-2010: a systematic analysis. Lancet Glob Health 2013;1:e339-49. [Crossref] [PubMed]
- Willis JR, Doan QV, Gleeson M, et al. Vision-Related Functional Burden of Diabetic Retinopathy Across Severity Levels in the United States. JAMA Ophthalmol 2017;135:926-32. [Crossref] [PubMed]
- NEI N. Diabetic Retinopathy Tables. 2019. Available online: https://www.nei.nih.gov/learn-about-eye-health/resources-for-health-educators/eye-health-data-and-statistics/diabetic-retinopathy-data-and-statistics/diabetic-retinopathy-tables. Accessed 1/12/2020.
- Cheng R, Ma JX. Angiogenesis in diabetes and obesity. Rev Endocr Metab Disord 2015;16:67-75. [Crossref] [PubMed]
- Busch C, Fraser-Bell S, Iglicki M, et al. Real-world outcomes of non-responding diabetic macular edema treated with continued anti-VEGF therapy versus early switch to dexamethasone implant: 2-year results. Acta Diabetol 2019;56:1341-50. [Crossref] [PubMed]
- Shah AR, Yonekawa Y, Todorich B, et al. Prediction of Anti-VEGF Response in Diabetic Macular Edema After 1 Injection. J Vitreoretin Dis 2017;1:169-74. [Crossref] [PubMed]
- Chen YP, Wu AL, Chuang CC, et al. Factors influencing clinical outcomes in patients with diabetic macular edema treated with intravitreal ranibizumab: comparison between responder and non-responder cases. Sci Rep 2019;9:10952. [Crossref] [PubMed]
- Brase JC, Wuttig D, Kuner R, et al. Serum microRNAs as non-invasive biomarkers for cancer. Mol Cancer 2010;9:306. [Crossref] [PubMed]
- Wittmann J, Jäck HM. Serum microRNAs as powerful cancer biomarkers. Biochim Biophys Acta 2010;1806:200-7. [PubMed]
- Fasanaro P, Greco S, Ivan M, et al. microRNA: emerging therapeutic targets in acute ischemic diseases. Pharmacol Ther 2010;125:92-104. [Crossref] [PubMed]
- Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A 2011;108:5003-8. [Crossref] [PubMed]
- Olivieri F, Rippo MR, Monsurrò V, et al. MicroRNAs linking inflamm-aging, cellular senescence and cancer. Ageing Res Rev 2013;12:1056-68. [Crossref] [PubMed]
- Olivieri F, Rippo MR, Procopio AD, et al. Circulating inflamma-miRs in aging and age-related diseases. Front Genet 2013;4:121. [Crossref] [PubMed]
- Somel M, Guo S, Fu N, et al. MicroRNA, mRNA, and protein expression link development and aging in human and macaque brain. Genome Res 2010;20:1207-18. [Crossref] [PubMed]
- Tahamtan A, Teymoori-Rad M, Nakstad B, et al. Anti-Inflammatory MicroRNAs and Their Potential for Inflammatory Diseases Treatment. Front Immunol 2018;9:1377. [Crossref] [PubMed]
- Liu CH, Huang S, Britton WR, et al. MicroRNAs in Vascular Eye Diseases. Int J Mol Sci 2020;21:649. [Crossref] [PubMed]
- Dexheimer PJ, Cochella L. MicroRNAs: From Mechanism to Organism. Front Cell Dev Biol 2020;8:409. [Crossref] [PubMed]
- McElhinney JMWR, Hasan A, Sajini AA. The epitranscriptome landscape of small noncoding RNAs in stem cells. Stem Cells 2020;38:1216-28. [Crossref] [PubMed]
- Roy D, Modi A, Khokhar M, et al. MicroRNA 21 Emerging Role in Diabetic Complications: A Critical Update. Curr Diabetes Rev 2021;17:122-35. [Crossref] [PubMed]
- Liu Y, Xiao J, Zhao Y, et al. microRNA-216a protects against human retinal microvascular endothelial cell injury in diabetic retinopathy by suppressing the NOS2/JAK/STAT axis. Exp Mol Pathol 2020;115:104445 [Crossref] [PubMed]
- Zeng Y, Cui Z, Liu J, et al. MicroRNA-29b-3p Promotes Human Retinal Microvascular Endothelial Cell Apoptosis via Blocking SIRT1 in Diabetic Retinopathy. Front Physiol 2019;10:1621. [Crossref] [PubMed]
- Han N, Xu H, Yu N, et al. MiR-203a-3p inhibits retinal angiogenesis and alleviates proliferative diabetic retinopathy in oxygen-induced retinopathy (OIR) rat model via targeting VEGFA and HIF-1α. Clin Exp Pharmacol Physiol 2020;47:85-94. [Crossref] [PubMed]
- Zhang ZZ, Qin XH, Zhang J. MicroRNA-183 inhibition exerts suppressive effects on diabetic retinopathy by inactivating BTG1-mediated PI3K/Akt/VEGF signaling pathway. Am J Physiol Endocrinol Metab 2019;316:E1050-60. [Crossref] [PubMed]
- Han N, Tian W, Yu N, et al. YAP1 is required for the angiogenesis in retinal microvascular endothelial cells via the inhibition of MALAT1-mediated miR-200b-3p in high glucose-induced diabetic retinopathy. J Cell Physiol 2020;235:1309-20. [Crossref] [PubMed]
- McArthur K, Feng B, Wu Y, et al. MicroRNA-200b regulates vascular endothelial growth factor-mediated alterations in diabetic retinopathy. Diabetes 2011;60:1314-23. [Crossref] [PubMed]
- Elfaki I, Mir R, Mir MM, et al. Potential Impact of MicroRNA Gene Polymorphisms in the Pathogenesis of Diabetes and Atherosclerotic Cardiovascular Disease. J Pers Med 2019;9:51. [Crossref] [PubMed]
- Zhang Y, Bai R, Liu C, et al. MicroRNA single-nucleotide polymorphisms and diabetes mellitus: A comprehensive review. Clin Genet 2019;95:451-61. [PubMed]
- Kaidonis G, Gillies MC, Abhary S, et al. A single-nucleotide polymorphism in the MicroRNA-146a gene is associated with diabetic nephropathy and sight-threatening diabetic retinopathy in Caucasian patients. Acta Diabetol 2016;53:643-50. [Crossref] [PubMed]
- McAuley AK, Dirani M, Wang JJ, et al. A genetic variant regulating miR-126 is associated with sight threatening diabetic retinopathy. Diab Vasc Dis Res 2015;12:133-8. [Crossref] [PubMed]
- Liu E, Kaidonis G, McComish BJ, et al. MicroRNA-Related Genetic Variants Are Associated With Diabetic Retinopathy in Type 1 Diabetes Mellitus. Invest Ophthalmol Vis Sci 2019;60:3937-42. [Crossref] [PubMed]
- Jiang C, Qin B, Liu G, et al. MicroRNA-184 promotes differentiation of the retinal pigment epithelium by targeting the AKT2/mTOR signaling pathway. Oncotarget 2016;7:52340-53. [Crossref] [PubMed]
- Liu CH, Wang Z, Sun Y, et al. Retinal expression of small non-coding RNAs in a murine model of proliferative retinopathy. Sci Rep 2016;6:33947. [Crossref] [PubMed]
- Polina ER, Oliveira FM, Sbruzzi RC, et al. Gene polymorphism and plasma levels of miR-155 in diabetic retinopathy. Endocr Connect 2019;8:1591-9. [Crossref] [PubMed]
- Zampetaki A, Kiechl S, Drozdov I, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res 2010;107:810-7. [Crossref] [PubMed]
- Jiménez-Lucena R, Camargo A, Alcalá-Diaz JF, et al. A plasma circulating miRNAs profile predicts type 2 diabetes mellitus and prediabetes: from the CORDIOPREV study. Exp Mol Med 2018;50:1-12. [Crossref] [PubMed]
- Jiménez-Lucena R, Rangel-Zúñiga OA, Alcalá-Díaz JF, et al. Circulating miRNAs as Predictive Biomarkers of Type 2 Diabetes Mellitus Development in Coronary Heart Disease Patients from the CORDIOPREV Study. Mol Ther Nucleic Acids 2018;12:146-57. [Crossref] [PubMed]
- Barutta F, Bellini S, Mastrocola R, et al. MicroRNA and Microvascular Complications of Diabetes. Int J Endocrinol 2018;2018:6890501 [Crossref] [PubMed]
- Qing S, Yuan S, Yun C, et al. Serum miRNA biomarkers serve as a fingerprint for proliferative diabetic retinopathy. Cell Physiol Biochem 2014;34:1733-40. [Crossref] [PubMed]
- Yin C, Lin X, Sun Y, et al. Dysregulation of miR-210 is involved in the development of diabetic retinopathy and serves a regulatory role in retinal vascular endothelial cell proliferation. Eur J Med Res 2020;25:20. [Crossref] [PubMed]
- Zampetaki A, Willeit P, Burr S, et al. Angiogenic microRNAs Linked to Incidence and Progression of Diabetic Retinopathy in Type 1 Diabetes. Diabetes 2016;65:216-27. [PubMed]
- Liu HN, Li X, Wu N, et al. Serum microRNA-221 as a biomarker for diabetic retinopathy in patients associated with type 2 diabetes. Int J Ophthalmol 2018;11:1889-94. [PubMed]
- Fiorentino L, Cavalera M, Mavilio M, et al. Regulation of TIMP3 in diabetic nephropathy: a role for microRNAs. Acta Diabetol 2013;50:965-9. [Crossref] [PubMed]
- Costantino S, Paneni F, Lüscher TF, et al. MicroRNA profiling unveils hyperglycaemic memory in the diabetic heart. Eur Heart J 2016;37:572-6. [Crossref] [PubMed]
- Lightell DJ Jr, Moss SC, Woods TC. Upregulation of miR-221 and -222 in response to increased extracellular signal-regulated kinases 1/2 activity exacerbates neointimal hyperplasia in diabetes mellitus. Atherosclerosis 2018;269:71-8. [Crossref] [PubMed]
- Liang Z, Gao KP, Wang YX, et al. RNA sequencing identified specific circulating miRNA biomarkers for early detection of diabetes retinopathy. Am J Physiol Endocrinol Metab 2018;315:E374-85. [Crossref] [PubMed]
- Martinez B, Peplow PV. MicroRNAs as biomarkers of diabetic retinopathy and disease progression. Neural Regen Res 2019;14:1858-69. [Crossref] [PubMed]
- Mammadzada P, Bayle J, Gudmundsson J, et al. Identification of Diagnostic and Prognostic microRNAs for Recurrent Vitreous Hemorrhage in Patients with Proliferative Diabetic Retinopathy. J Clin Med 2019;8:2217. [Crossref] [PubMed]
- Chen S, Yuan M, Liu Y, et al. Landscape of microRNA in the aqueous humour of proliferative diabetic retinopathy as assessed by next-generation sequencing. Clin Exp Ophthalmol 2019;47:925-36. [Crossref] [PubMed]
- Smit-McBride Z, Nguyen KNH, Lai AW, et al. The Effect of DR biomarkers microRNAs on VEGF Secretion in human retinal pigment epithelial cells.ARVO; April 29 - May 3, 2018; Honolulu, Hawaii 2018. Available online: https://iovs.arvojournals.org/article.aspx?articleid=2692747
- Smit-McBride Z, Chen YY, Bui MC, et al., editors. Effect of DR microRNA biomarkers on angiogenesis assay activity in vitro. ARVO; 2019; Vancouver, Canada. Available online: https://iovs.arvojournals.org/article.aspx?articleid=2742711
- Smit-McBride Z, Okafor K, Nguyen AT, et al. Comparative expression analysis in plasma of circulating microRNAs identified in ocular fluids as putative biomarkers for diabetic retinopathy ARVO; Denver, Colorado 2015. Available online: https://iovs.arvojournals.org/article.aspx?articleid=2335191
- Smit-McBride Z, Nguyen AT, Yu AK, et al. Unique molecular signatures of microRNAs in ocular fluids and plasma in diabetic retinopathy. PLoS One 2020;15:e0235541 [Crossref] [PubMed]
- Li Z, Dong Y, He C, et al. RNA-Seq Revealed Novel Non-proliferative Retinopathy Specific Circulating MiRNAs in T2DM Patients. Front Genet 2019;10:531. [Crossref] [PubMed]
- Zhou H, Peng C, Huang DS, et al. microRNA Expression Profiling Based on Microarray Approach in Human Diabetic Retinopathy: A Systematic Review and Meta-Analysis. DNA Cell Biol 2020;39:441-50. [Crossref] [PubMed]
- Frost RJ, Olson EN. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci U S A 2011;108:21075-80. [Crossref] [PubMed]
- Gong Q, Su G. Roles of miRNAs and long noncoding RNAs in the progression of diabetic retinopathy. Biosci Rep 2017;37:BSR20171157 [Crossref] [PubMed]
- Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol 2011;21:354-61. [Crossref] [PubMed]
- Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018;172:393-407. [Crossref] [PubMed]
- Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet 2016;17:272-83. [Crossref] [PubMed]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell 2011;43:904-14. [Crossref] [PubMed]
- Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. RNA 2010;16:2043-50. [Crossref] [PubMed]
- Yan B, Tao ZF, Li XM, et al. Aberrant expression of long noncoding RNAs in early diabetic retinopathy. Invest Ophthalmol Vis Sci 2014;55:941-51. [Crossref] [PubMed]
- Qiu GZ, Tian W, Fu HT, et al. Long noncoding RNA-MEG3 is involved in diabetes mellitus-related microvascular dysfunction. Biochem Biophys Res Commun 2016;471:135-41. [Crossref] [PubMed]
- Zhang J, Chen M, Chen J, et al. Long non-coding RNA MIAT acts as a biomarker in diabetic retinopathy by absorbing miR-29b and regulating cell apoptosis. Biosci Rep 2017;37:BSR20170036 [Crossref] [PubMed]
- Yan B, Yao J, Liu JY, et al. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ Res 2015;116:1143-56. [Crossref] [PubMed]
- Sun Y, Liu YX. LncRNA HOTTIP improves diabetic retinopathy by regulating the p38-MAPK pathway. Eur Rev Med Pharmacol Sci 2018;22:2941-8. [PubMed]
- Biswas S, Thomas AA, Chen S, et al. MALAT1: An Epigenetic Regulator of Inflammation in Diabetic Retinopathy. Sci Rep 2018;8:6526. [Crossref] [PubMed]
- Liu JY, Yao J, Li XM, et al. Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis 2014;5:e1506 [Crossref] [PubMed]
- Li CP, Wang SH, Wang WQ, et al. Long Noncoding RNA-Sox2OT Knockdown Alleviates Diabetes Mellitus-Induced Retinal Ganglion Cell (RGC) injury. Cell Mol Neurobiol 2017;37:361-9. [Crossref] [PubMed]
- Thomas AA, Feng B, Chakrabarti S. ANRIL: A Regulator of VEGF in Diabetic Retinopathy. Invest Ophthalmol Vis Sci 2017;58:470-80. [Crossref] [PubMed]
- Li Y, Xu F, Xiao H, et al. Long noncoding RNA BDNF-AS inversely regulated BDNF and modulated high-glucose induced apoptosis in human retinal pigment epithelial cells. J Cell Biochem 2018;119:817-23. [Crossref] [PubMed]
- Shao J, Zhang Y, Fan G, et al. Transcriptome analysis identified a novel 3-LncRNA regulatory network of transthyretin attenuating glucose induced hRECs dysfunction in diabetic retinopathy. BMC Med Genomics 2019;12:134. [Crossref] [PubMed]
- Thomas AA, Biswas S, Feng B, et al. lncRNA H19 prevents endothelial-mesenchymal transition in diabetic retinopathy. Diabetologia 2019;62:517-30. [Crossref] [PubMed]
- Yu L, Fu J, Yu N, et al. Long noncoding RNA MALAT1 participates in the pathological angiogenesis of diabetic retinopathy in an oxygen-induced retinopathy mouse model by sponging miR-203a-3p. Can J Physiol Pharmacol 2020;98:219-27. [Crossref] [PubMed]
- Liu XQ, Duan LS, Chen YQ, et al. lncRNA MALAT1 Accelerates Wound Healing of Diabetic Mice Transfused with Modified Autologous Blood via the HIF-1α Signaling Pathway. Mol Ther Nucleic Acids 2019;17:504-15. [Crossref] [PubMed]
- Li ZX, Zhu QN, Zhang HB, et al. MALAT1: a potential biomarker in cancer. Cancer Manag Res 2018;10:6757-68. [Crossref] [PubMed]
- Shaker OG, Abdelaleem OO, Mahmoud RH, et al. Diagnostic and prognostic role of serum miR-20b, miR-17-3p, HOTAIR, and MALAT1 in diabetic retinopathy. IUBMB Life 2019;71:310-20. [Crossref] [PubMed]
- Boon RA, Jaé N, Holdt L, et al. Long Noncoding RNAs: From Clinical Genetics to Therapeutic Targets? J Am Coll Cardiol 2016;67:1214-26. [Crossref] [PubMed]
- Li Q, Pang L, Yang W, et al. Long Non-Coding RNA of Myocardial Infarction Associated Transcript (LncRNA-MIAT) Promotes Diabetic Retinopathy by Upregulating Transforming Growth Factor-β1 (TGF-β1) Signaling. Med Sci Monit 2018;24:9497-503. [Crossref] [PubMed]
- Battegay EJ. Angiogenesis: mechanistic insights, neovascular diseases, and therapeutic prospects. J Mol Med (Berl) 1995;73:333-46. [Crossref] [PubMed]
- Nunes I, Munger J, Harpel JG, et al. Structure and activation of the large latent transforming growth factor-Beta complex. J Am Optom Assoc 1998;69:643-8. [PubMed]
- Pfeiffer A, Spranger J, Meyer-Schwickerath R, et al. Growth factor alterations in advanced diabetic retinopathy: a possible role of blood retina barrier breakdown. Diabetes 1997;46:S26-30. [Crossref] [PubMed]
- Vidal L, Blagden S, Attard G, et al. Making sense of antisense. Eur J Cancer 2005;41:2812-8. [Crossref] [PubMed]
- Christopher AF, Kaur RP, Kaur G, et al. MicroRNA therapeutics: Discovering novel targets and developing specific therapy. Perspect Clin Res 2016;7:68-74. [Crossref] [PubMed]
- Ratti M, Lampis A, Ghidini M, et al. MicroRNAs (miRNAs) and Long Non-Coding RNAs (lncRNAs) as New Tools for Cancer Therapy: First Steps from Bench to Bedside. Target Oncol 2020;15:261-78. [Crossref] [PubMed]
- Elmén J, Lindow M, Schütz S, et al. LNA-mediated microRNA silencing in non-human primates. Nature 2008;452:896-9. [Crossref] [PubMed]
- Veedu RN, Wengel J. Locked nucleic acids: promising nucleic acid analogs for therapeutic applications. Chem Biodivers 2010;7:536-42. [Crossref] [PubMed]
- Leucci E, Vendramin R, Spinazzi M, et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature 2016;531:518-22. [Crossref] [PubMed]
- Michalik KM, You X, Manavski Y, et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res 2014;114:1389-97. [Crossref] [PubMed]
- Viereck J, Kumarswamy R, Foinquinos A, et al. Long noncoding RNA Chast promotes cardiac remodeling. Sci Transl Med 2016;8:326ra22 [Crossref] [PubMed]
- Xing Z, Lin A, Li C, et al. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell 2014;159:1110-25. [Crossref] [PubMed]
- Li L, Zhu D, Huang L, et al. Argonaute 2 complexes selectively protect the circulating microRNAs in cell-secreted microvesicles. PLoS One 2012;7:e46957 [Crossref] [PubMed]
- György B, Hung ME, Breakefield XO, et al. Therapeutic applications of extracellular vesicles: clinical promise and open questions. Annu Rev Pharmacol Toxicol 2015;55:439-64. [Crossref] [PubMed]
- György B, Szabó TG, Pásztói M, et al. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell Mol Life Sci 2011;68:2667-88. [Crossref] [PubMed]
- Ståhl AL, Johansson K, Mossberg M, et al. Exosomes and microvesicles in normal physiology, pathophysiology, and renal diseases. Pediatr Nephrol 2019;34:11-30. [Crossref] [PubMed]
- Stahl PD, Raposo G. Extracellular Vesicles: Exosomes and Microvesicles, Integrators of Homeostasis. Physiology (Bethesda) 2019;34:169-77. [Crossref] [PubMed]
- Das CK, Jena BC, Banerjee I, et al. Exosome as a Novel Shuttle for Delivery of Therapeutics across Biological Barriers. Mol Pharm 2019;16:24-40. [Crossref] [PubMed]


