
MiR-495 regulates cell proliferation and apoptosis in H2O2stimulated rat spinal cord neurons through targeting signal transducer and activator of transcription 3 (STAT3)
Introduction
Spinal cord injury (SCI) is a debilitating condition that contributes to post-traumatic tissue functional deficits (1). Initial damage to the spinal cord may simultaneously disintegrate cell membranes, injure myelin and axons, and cause damage to micro-vessels, thereby activating worse secondary injuries (2). The secondary injury cascades are functional biological processes which include local inflammation, free radical production, and increased oxidative stress, all of which may induce neuronal apoptosis, glial cell death and axonal destruction, ultimately leading to permanent neurological disability (3,4). Previous studies have demonstrated that hypoxia (5), ischemia (6), lipid peroxidation (7), and ROS (7) are implicated in the underlying pathophysiology of SCI-induced neuronal apoptosis. Interestingly, hydrogen peroxide (H2O2) induces neuronal death in the spinal cord through intracellular ROS generation and the modulation of genes related to apoptosis (8). Moreover, the microRNA-21 (miR-21) has been shown to regulate cell proliferation and apoptosis in H2O2-stimulated neurons of the spinal cord, and it controlled ROS generation by its effect on Smad7 in mouse SCNs (9). Although the internal and external signals modulating the apoptosis of neurons have been much explored, the mechanisms that underlie the effect of miRNAs on neuronal apoptosis following SCI are still unclear.
MicroRNAs, a category of endogenous molecules (18-25 nucleotides) which do not code for amino acids, exert regulatory influence on transcription via interaction with the 3'-UTRs of mRNA targets (10). MiRNAs play important roles in cell proliferation, differentiation, and apoptosis in various cell types (11,12). Several studies have found that they are associated with several pathological tissue lesions, as well as uncommon miRNA expressions, although various cell signaling pathways regulate the development of neurons (13,14). MiR-93, by targeting Eph-A4, has been shown to increase neurite formation from neurons of the spinal cord (15). Furthermore, the up-regulation of miR-26a promoted neurite growth, and also protected the spinal cord against local analgesia-induced nerve damage (16). MiR-9 has been shown to modulate the apoptosis of neurons by targeting monocyte chemotactic protein-induced protein-1 expression in a mouse model of SCI (17). MiR-124 is highly expressed in neurons and has important functions in embryonic neurogenesis and postnatal neuronal differentiation. However, miR-124 expression in neurons was down-regulated after SCI in a mouse model (18). These results indicate that miRNAs are the upstream molecular regulators of neural differentiation, and may be potential therapeutic targets for SCI. Importantly, many miRNAs have been shown to be markedly up-regulated or down-regulated in H2O2-stimulated rat SCNs (9). However, the molecular mechanisms involved in the effect of miR-459 on SCI, and the involvement of STAT3 remain unknown.
In the present study, H2O2 was added to cell cultures so as to induce oxidative stress, a phenomenon related to secondary SCI, and we hypothesized that microRNA-495 (miR-495) may regulate STAT3 expression and its involvement in responses to H2O2-stimulated rat SCNs. We present the following article in accordance with the MDAR reporting checklist (http://dx.doi.org/10.21037/atm-21-102).
Methods
Cultural conditions
Primary neurons of the spinal cord were harvested from embryonic day 14 Sprague-Dawley rats as previously described (19). The cells were cultured at 37 °C in high-glucose DMEM containing 10% heat-inactivated FBS and 5% heat-inactivated horse serum (Gibco Lab) in a humidified atmosphere containing 5% CO2 and 95% O2.
MTT assay
To assess cell viability, the MTT assay was performed on SCN outgrowths using an assay kit (Shanghai Jining Industrial Co., LTD., China) in line with the instructions on the kit manual.
Measurement of caspase 3 activity
SCNs were lysed and then stained using anti-caspase-3 (Shanghai Enzyme-linked Biotechnology Co., Ltd. China). Immunocomplexes were formed after 2 hours, and the absorbance of the p-nitroaniline liberated was read at 405 nm in an ELISA instrument.
TUNEL assay
Apoptosis was measured using terminal dUTP nick-end labeling (TUNEL) assay. Following trypsinization, SCNs were exposed to 4% paraformaldehyde, and subjected to permeabilization with Triton-X-100 in 0.1% Na citrate. After washing, cells were put in an incubator along with the reaction mixture for 60 minutes at 37 °C, after which they were immediately analyzed using FACScan (Becton Dickinson) and the Cellquest program.
Lactate dehydrogenase (LDH) activity assay
SCNs were seeded in 96-well plates, and then treated with H2O2. After 24 hours, cells were centrifuged to obtain the supernatants which were subjected to LDH assay using assay kits (Keygen, Nanjing, China) in line with manual protocol. Data were normalized with the concentration of podocyte protein lysates.
ROS measurements
Intracellular ROS generation was determined by measuring the uptake of DCFH-DA. The SCNs were seeded in well plates and treated with the dye for 24 hours. Then, the cells were analyzed using flow cytometry (Becton Dickinson).
Comet assay
Frozen slides were fully precoated with 0.8% agarose in PBS of pH approximately 7.4, and also blanketed with 22 mm × 22 mm glass coverslips, and kept for 20 min at laboratory temperature. Next, a 3:7 volume ratio of cell culture and 1% low MP agarose was evenly spread over every corner of the precoated slides, blanketed, and imaged with an Olympus fluorescence microscope bearing a CCD camera. Comet tail lengths were calculated in every group with Image proR plus appliance. Olive tail moment (OTM) denoted DNA damage, and it was quantified viz:
Luciferase reporter assay
The 3'-untranslated regions of the STAT3 gene bearing the predicted target points for miR-495 were got via polymerase chain reaction amplification. The construct was placed into several cloning points in pMIR-REPORT luciferase miRNA reporter vector. SCNs were subjected to co-transfection with Lipofectamine 2000 using 0.1 µg of luciferase reporters bearing miR-495 mimics and STAT3 3'-UTR. Cell lysates were obtained 48 hours after transfection, and luciferase was measured using the dual luciferase reporter assay kits.
Transfection of miR-495 inhibitors and mimics
FAM-derivatized 2'-OMe-oligonucleotides were produced by GenePharma, Shanghai, China. The sequence of 2'-OMe miR-495 mimic which comprised RNA duplexes was: 5'-AAACAAACAUGGUGCACUUCUU-3, while 2'-OMe miR-495 inhibitor and 2'-OMe scramble oligonucleotides had sequences of 5'-AAGAAGUGCACCAUGUUUGUUU-3' and 5'-ACUUGCGUUAUUGGUGACCUAC-3', respectively. Transfection was done with Lipofectamine 2000, with change of medium after 24 hours and cell analysis after 48 hours.
RT-Polymerase chain reaction
Total RNA was extracted with TRIzol. Then, 4 µg of RNA was reverse-transcribed to cDNA using MMLV reverse transcriptase in addition to oligo(dT) primers (Fermentas) in line with the kit protocols. MiR-495 was quantified using mirVana qRT-PCR miRNA detection kit (Ambion, Austin, USA) with SYBR Green. The following primers were used:
STAT3: 5'-GGGTGGAGAAGGACATCAGCGGTAA-3' (sense); 5'-GCCGACAATACTTTCCGAATCC-3' (anti-sense);
GAPDH: 5'-GCACCGTCAAGCTGAGAAC-3' (sense); 5'-TGGTGAAGACGCCAGTGGA-3' (antisense).
The relative mRNA expression was calculated using the 2-ΔΔCT method.
Immunoblot assay
SCNs were lysed using Nonidet P-40 Lysis buffer, boiled for e few minutes, and then centrifuged to obtain the supernatants. Solutions with 50 µg protein were subjected to 10% SDS-polyacrylamide gel electrophoresis, followed by transfer onto NC membranes which were then blocked with 5% (w/v) fat-free milk powder prior to incubation for 12 hours at 4 °C with 10 antibodies for Bcl-2, Bax, caspase 3, and STAT3. Following rinsing with TBST, incubation with horse radish peroxidase-conjugated-antibody was carried out at room temperature for 2 hours, followed by visualization with ECL.
Statistical analysis
Data are shown as mean ± SEM, and were analyzed using analysis of variance and Tukey’s test. GraphPad Prism version 6 was employed analysis. Statistical significance was assumed at P<0.05.
Results
H2O2-induced cytotoxicity and apoptosis in SCNs
The cytotoxicity and apoptosis of SCNs were assessed after exposure to different concentrations of H2O2. The H2O2 exposure decreased the viability of SCNs in a dose- and time-dependent manner (Figure 1A). The mean lethal concentration of H2O2 in SCNs after 24 hours incubation was approximately 40 µM (Figure 1A). Moreover, we examined H2O2-induced apoptosis in SCNs using caspase-3 levels and TUNEL staining 24 hours after H2O2 incubation. As shown in Figures 1B,C, H2O2 exposure induced cell apoptosis and increased the levels of caspase 3 in SCN lysates in a dose-reliant fashion. Furthermore, protein expressions of markers relating to apoptosis were measured using western blot assay. H2O2 treatment resulted in down-regulated expression of anti-apoptotic Bcl-2 (Figure 1D), and up-regulated expressions Bax (Figure 1E) and caspase-3 (Figure 1F) in SCNs.
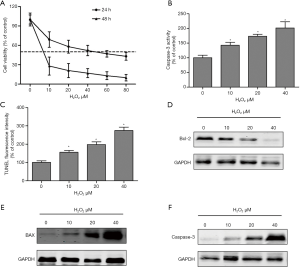
H2O2-induced cell membrane dysfunction, ROS production, and DNA damage
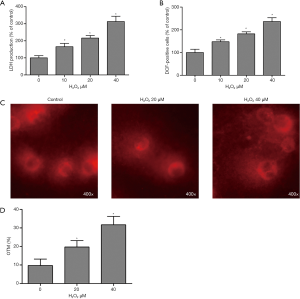
A concentration-dependent increase in extracellular LDH activity was observed in SCNs exposed to H2O2 at concentrations greater than 10 µM (Figure 2A). Moreover, the results indicated that H2O2 exposure led to marked rise in ROS levels in a concentration-dependent manner (Figure 2B). The comet assay was performed to assess the genotoxicity of H2O2 in SCNs. As shown in Figures 2C,D, when the SCNs were treated with H2O2, olive tail moment was markedly higher than that in the control group.
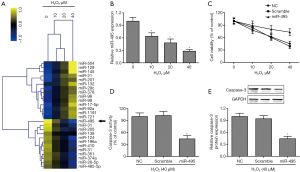
Overexpressed miR-495 inhibited H2O2-induced SCN apoptosis
To identify the differentially expressed miRNAs in cultured SCNs in response to H2O2 exposure, we performed a microarray assay with miRNA libraries generated using total RNA extracted from H2O2-stimulated (0, 10, 20, or 40 µM) SCNs for 24 hours. We found that miR-495 was significantly lowly expressed in H2O2-treated SCNs, relative to control (Figure 3A). Therefore, we further investigated the function of miR-495 in SCNs when they were exposed to H2O2. The dose-dependent experiments showed that H2O2 markedly suppressed miR-495 expression in SCNs (Figure 3B). However, miR-495 gain-of-function reversed H2O2-induced cytotoxicity (Figure 3C) and apoptosis (Figure 3D,E) in SCNs. Moreover, we found that H2O2-treated SCNs had up-regulated mRNA (Figure 4A) and protein (Figure 4B) expressions of STAT3.
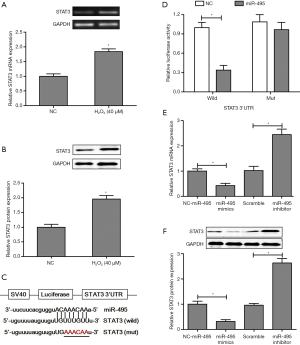
Based on the miRBase database (http://www.mirbase.org), we found the hypothetical interaction domain of miR-495 in 3'-UTR of STAT3 in mice (Figure 4C). To verify whether STAT3 was a direct target of miR-495, the 3'-UTR of the wild-type and mutant STAT3 genes were cloned and co-transfected along with miR-495 or NC oligonucleotides into SCNs. Luciferase assays were performed 24 hours post-transfection. As shown in Figure 4D, there was reduced activity of the enzyme in miR-495-transfected SCNs, when compared to NC cells. In contrast, after co-transfection of miR-495 into STAT3 mutant 3'-UTR cells, the activity of luciferase was not significantly different from that of the NC. Next, we observed that transfection of SCNs with miR-495 mimic oligonucleotides or miR-495 inhibitors significantly down-regulated or up-regulated both STAT3 mRNA (Figure 4E) and protein (Figure 4F) expression levels, respectively. Thus, miR-495 exerted a mitigating influence against H2O2-induced SCNs by suppressing the expression of STAT3.
Discussion
Our data suggest that miR-495 was involved in H2O2-induced SCN dysfunction. Moreover, H2O2 induced apoptosis, ROS, and DNA damage in SCNs. Simultaneously, H2O2 exposure down-regulated miR-495 levels and up-regulated STAT3 levels in SCNs. Interestingly, overexpressed miR-495 reversed H2O2-induced cytotoxicity and apoptosis in SCNs, and inhibited the mRNA and protein expression of STAT3. Therefore, we conclude that miR-495 may exert a protective effect against H2O2-induced SCN dysfunction by suppressing the expression of STAT3.
MiRNAs regulate translation by interacting with the 3'-UTR of target mRNAs, thereby suppressing protein translation in cells (20,21). In this study, we found potential miR-495 binding domains within 3'-UTR of STAT3 in rats. In addition, miR-495 regulated luciferase expression by interacting with the 3'-UTR of STAT3 in SCNs. Moreover, our results indicated that STAT3 expression was suppressed by miR-495 overexpression at the transcriptional and translational levels. MiR-495 is involved in the pathogenesis of several types of cancers (22,23). However, to our knowledge, there are no extant studies on the neuroprotective effect of miR-495 against SCI. In response to H2O2-stimulated SCN dysfunction in rats, miR-495 had the lowest expression in the presence of H2O2. However, overexpressed miR-495 reversed H2O2-induced cytotoxicity and apoptosis in SCNs. Importantly, we addressed the post-translational phenomenon of miR-495 and the possible targets of miR-495 during neuronal apoptosis following SCI. Bioinformatics analysis showed the potential miR-495 binding domains in 3'-UTR of STAT3 in rat SCNs.
Previous studies have indicated that STAT3 is a key regulator of astrocytes (which are reactive in nature) during the repair process after SCI. Therefore, it is an important target for the treatment of CNS injury (24,25). It has been reported that activation of STAT3 route may be implicated in the development of neuropathic pain in rats (26). Moreover, the activation of STAT3 is involved in IL-17-induced spinal cord neuroinflammation after SCI in rats (27). In contrast, sustained activation of STAT3 has been shown to enhance corticospinal remodeling and functional recovery after SCI (28). This investigation has shown that H2O2 treatment up-regulated mRNA and protein expressions of STAT3 in SCNs. STAT3, a widely studied transcription factor which is known to be involved in the protection of neurons and nerve regeneration, exists in cytosol in a sedentary state. However, peripheral damage of the nerve increases the transcription STAT3 (29). In a mouse model of hypoglossal nerve damage, miR-124 was shown to be implicated in the regulation of the expression of STAT3, which was crucial for proper nerve regeneration (30). In the present study, we cloned the 3'-UTR of the wild-type or mutant-type STAT3 gene and co-transfected it along with miR-495 or scramble sequences into SCNs, which resulted in reduced luciferase activity in miR-495-transfected cells. These results showed that miR-495 was capable of suppressing endogenous STAT3 expression via targeting its 3'-UTRs.
In summary, this study has indicated that miR-495 suppresses STAT3 in SCNs. Overexpression of miR-495 might play a protective role against H2O2-induced SCN dysfunction by suppressing the expression of STAT3.
Acknowledgments
Funding: This work was supported by the Science and Technology Commission of Jiading District, Shanghai (JDKW-2018-W03).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-102
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-102
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-102). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- DePaul MA, Palmer M, Lang BT, et al. Intravenous multipotent adult progenitor cell treatment decreases inflammation leading to functional recovery following spinal cord injury. Sci Rep 2015;5:16795. [Crossref] [PubMed]
- Kwon BK, Tetzlaff W, Grauer JN, et al. Pathophysiology and pharmacologic treatment of acute spinal cord injury. Spine J 2004;4:451-64. [Crossref] [PubMed]
- Elkabes S, Nicot AB. Sex steroids and neuroprotection in spinal cord injury: a review of preclinical investigations. Exp Neurol 2014;259:28-37. [Crossref] [PubMed]
- Wu J, Kharebava G, Piao C, et al. Inhibition of E2F1/CDK1 pathway attenuates neuronal apoptosis in vitro and confers neuroprotection after spinal cord injury in vivo. PLoS One 2012;7:e42129 [Crossref] [PubMed]
- Lee KZ, Sandhu MS, Dougherty BJ, et al. Hypoxia triggers short term potentiation of phrenic motoneuron discharge after chronic cervical spinal cord injury. Exp Neurol 2015;263:314-24. [Crossref] [PubMed]
- Li Y, Gu J, Liu Y, et al. iNOS participates in apoptosis of spinal cord neurons via p-BAD dephosphorylation following ischemia/reperfusion (I/R) injury in rat spinal cord. Neurosci Lett 2013;545:117-22. [Crossref] [PubMed]
- Hassler SN, Johnson KM, Hulsebosch CE. Reactive oxygen species and lipid peroxidation inhibitors reduce mechanical sensitivity in a chronic neuropathic pain model of spinal cord injury in rats. J Neurochem 2014;131:413-7. [Crossref] [PubMed]
- Jiang X, Nie B, Fu S, et al. EGb761 protects hydrogen peroxide-induced death of spinal cord neurons through inhibition of intracellular ROS production and modulation of apoptotic regulating genes. J Mol Neurosci 2009;38:103-13. [Crossref] [PubMed]
- Jiao G, Pan B, Zhou Z, et al. MicroRNA-21 regulates cell proliferation and apoptosis in H(2)O(2)-stimulated rat spinal cord neurons. Mol Med Rep 2015;12:7011-6. [Crossref] [PubMed]
- Paranjape T, Heneghan H, Lindner R, et al. A 3'-untranslated region KRAS variant and triple-negative breast cancer: a case-control and genetic analysis. Lancet Oncol 2011;12:377-86. [Crossref] [PubMed]
- Fujii T, Shimada K, Tatsumi Y, et al. microRNA-145 promotes differentiation in human urothelial carcinoma through down-regulation of syndecan-1. BMC Cancer 2015;15:818. [Crossref] [PubMed]
- Hata A, Lieberman J. Dysregulation of microRNA biogenesis and gene silencing in cancer. Sci Signal 2015;8:re3. [Crossref] [PubMed]
- Sathyan P, Zinn PO, Marisetty AL, et al. Mir-21-Sox2 Axis Delineates Glioblastoma Subtypes with Prognostic Impact. J Neurosci 2015;35:15097-112. [Crossref] [PubMed]
- Zhang W, Kim PJ, Chen Z, et al. MiRNA-128 regulates the proliferation and neurogenesis of neural precursors by targeting PCM1 in the developing cortex. Elife 2016;5:e11324 [Crossref] [PubMed]
- Chen X, Yang H, Zhou X, et al. MiR-93 Targeting EphA4 Promotes Neurite Outgrowth from Spinal Cord Neurons. J Mol Neurosci 2016;58:517-24. [Crossref] [PubMed]
- Cui C, Xu G, Qiu J, et al. Up-regulation of miR-26a promotes neurite outgrowth and ameliorates apoptosis by inhibiting PTEN in bupivacaine injured mouse dorsal root ganglia. Cell Biol Int 2015;39:933-42. [Crossref] [PubMed]
- Xu Y, An BY, Xi XB, et al. MicroRNA-9 controls apoptosis of neurons by targeting monocyte chemotactic protein-induced protein 1 expression in rat acute spinal cord injury model. Brain Res Bull 2016;121:233-40. [Crossref] [PubMed]
- Zhao Y, Zhang H, Zhang D, et al. Loss of microRNA-124 expression in neurons in the peri-lesion area in mice with spinal cord injury. Neural Regen Res 2015;10:1147-52. [Crossref] [PubMed]
- Jiang XY, Fu SL, Nie BM, et al. Methods for isolating highly-enriched embryonic spinal cord neurons: a comparison between enzymatic and mechanical dissociations. J Neurosci Methods 2006;158:13-8. [Crossref] [PubMed]
- Liu CH, Sun Y, Li J, et al. Endothelial microRNA-150 is an intrinsic suppressor of pathologic ocular neovascularization. Proc Natl Acad Sci U S A 2015;112:12163-8. [Crossref] [PubMed]
- Wu J, Zheng C, Wang X, et al. MicroRNA-30 family members regulate calcium/calcineurin signaling in podocytes. J Clin Invest 2015;125:4091-106. [Crossref] [PubMed]
- Hwang-Verslues WW, Chang PH, Wei PC, et al. miR-495 is upregulated by E12/E47 in breast cancer stem cells, and promotes oncogenesis and hypoxia resistance via downregulation of E-cadherin and REDD1. Oncogene 2011;30:2463-74. [Crossref] [PubMed]
- Li Z, Cao Y, Jie Z, et al. miR-495 and miR-551a inhibit the migration and invasion of human gastric cancer cells by directly interacting with PRL-3. Cancer Lett 2012;323:41-7. [Crossref] [PubMed]
- Okada S, Nakamura M, Katoh H, et al. Conditional ablation of Stat3 or Socs3 discloses a dual role for reactive astrocytes after spinal cord injury. Nat Med 2006;12:829-34. [Crossref] [PubMed]
- Herrmann JE, Imura T, Song B, et al. STAT3 is a critical regulator of astrogliosis and scar formation after spinal cord injury. J Neurosci 2008;28:7231-43. [Crossref] [PubMed]
- Dominguez E, Rivat C, Pommier B, et al. JAK/STAT3 pathway is activated in spinal cord microglia after peripheral nerve injury and contributes to neuropathic pain development in rat. J Neurochem 2008;107:50-60. [Crossref] [PubMed]
- Zong S, Zeng G, Fang Y, et al. The role of IL-17 promotes spinal cord neuroinflammation via activation of the transcription factor STAT3 after spinal cord injury in the rat. Mediators Inflamm 2014;2014:786947 [Crossref] [PubMed]
- Lang C, Bradley PM, Jacobi A, et al. STAT3 promotes corticospinal remodelling and functional recovery after spinal cord injury. EMBO Rep 2013;14:931-7. [Crossref] [PubMed]
- Qiu J, Cafferty WB, McMahon SB, et al. Conditioning injury-induced spinal axon regeneration requires signal transducer and activator of transcription 3 activation. J Neurosci 2005;25:1645-53. [Crossref] [PubMed]
- Nagata K, Hama I, Kiryu-Seo S, et al. microRNA-124 is down regulated in nerve-injured motor neurons and it potentially targets mRNAs for KLF6 and STAT3. Neuroscience 2014;256:426-32. [Crossref] [PubMed]
(English Language Editor: C. Betlazar-Maseh)


