Accuracy of matrix-assisted laser desorption ionization time-of-flight mass spectrometry for direct bacterial identification from culture-positive urine samples
Introduction
Urinary tract infection (UTI) is one of the leading causes of morbidity and mortality in primary care and hospital settings and is associated with hospital-acquired and catheter-associated infection (1). About 150 million people suffer from this disease, and total economic burden of treatment exceeds $6 billion worldwide per year (2). In recent years, molecular microbiology technologies, such as Biosensors, 16S RNA gene sequencing, and different PCR-based techniques, have been widely adopted by clinical laboratories to accelerate, simplify, and perform precise detection of specific pathogens involved in UTI. However, these methods are impractical for routine use due to their associated complicated procedures and high cost. Conventional phenotypic and biochemical methods, such as urine culture, are important components of clinical and public health laboratories. Urine culture is still regarded as the gold standard for diagnosis for UTI. This method cannot be replaced by any advanced methods in identifying specific urinary tract pathogens. Nevertheless, the main disadvantage of conventional methods is its long turnaround time (TAT). This approach takes nearly 18–36 h for final identification of positive samples, and antimicrobial susceptibility testing consumes an additional 18–24 h in conventional laboratory UTI diagnostics. Antibacterial resistance has significantly increased worldwide due to the inappropriate empirical antibiotic therapy of UTI (3). Therefore, a novel diagnostic methodology that could provide rapid and accurate information within few hours is needed for timely identification of pathogens in clinical settings.
Matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) has been recognized as a new standard for bacterial identification in clinical microbiology laboratories. MALDI-TOF MS is a powerful and high-throughput microbial identification technology that is efficient, rapid, relatively cheap, and easy to use. Compared with conventional methods, MALDI-TOF MS can reduce workload in clinical microbial identification and significantly shorten TAT before delivering the reports to clinicians. Studies have successfully reported that MALDI-TOF MS can identify bacteria directly from culture-positive urine samples. Nevertheless, the results vary because several limitations should be considered in rapid MALDI-TOF MS-based identification of clinical specimens. First, a sufficient concentration of bacterial cells is required to obtain a valid protein spectrum, which is critical to urine specimens (4). Second, the presence of human cells, such as leucocytes, erythrocytes, and other organic components, complicates the separation of microorganisms from human cells and therefore pre-treatment is required before analysis (5). The rate of negative samples can reach 60–70% of the total urine samples received (6,7). A practical method is needed to detect negative urine samples.
Scholars have reported a successful urine processing and preparation protocol for bacterial isolation and identification. The protocol mainly involves selection, washing, and centrifugation to obtain a pellet of bacteria for further identification (8-10). Each received urine sample is analyzed with a flow cytometry system or other instruments to screen bacterial counts. All samples with a cut-off are selected and further processed by MALDI-TOF MS for pathogen detection (8). The selected samples are vortexed and centrifuged to remove impurities, such as cellular debris or mucus. The resulting supernatant is then centrifuged to collect bacteria pellets (9). The pellets are resuspended and washed by centrifugation again. Finally, bacterial purification is accomplished (10).
This systematic review aims to evaluate the accuracy of MALDI-TOF MS for direct identification of monomicrobial urine specimens by searching related publications in literature. We analyzed variables that may influence the accuracy of bacterial identification at the species level.
We present this systematic literature review by referring to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (11) (available at http://dx.doi.org/10.21037/atm-20-7310).
Methods
Search strategy
We systematically queried the electronic database of Medline and Web of Science (before 1 May 2020) with the string “(MALDI-TOF [MeSH Terms] AND urine specimens/samples [MeSH Terms] OR urinary tract infections [MeSH Terms]) AND (identification [Title/Abstract] AND accuracy [Title/Abstract])” to identify relevant articles that assessed the accuracy of MALDI-TOF MS for direct identification of bacteria from clinical urine specimens. Publication status, geographical distribution, and language restrictions were not applied. Duplicate articles identified in Medline and Web of Science were manually deleted using Reference Manager (Thomson Reuters EndNote X9).
Study selection criteria
Eligible studies considered for systematic review were those that quantitatively evaluated the accuracy of MALDI-TOF MS (Bruker Daltonics, Bremen, Germany) for direct bacterial identification of clinical urine cultures in comparison with routine bacterial identification methods (phenotypic, microbiological, molecular diagnosis). Data or studies were excluded if they fell into the following categories: studies without a comparator method or identification standard; studies that did not investigate urine cultures; studies on technological innovations; studies with no abstract; non-original articles; non-human studies; and case-series studies, editorials, letters, or reviews.
Data extraction
Literature search and data extraction were arbitrated by two investigators (BS and WW), and the full paper of each potentially eligible study was obtained. Discrepancies were discussed, and the opinion of a third investigator (BG) was sought when necessary to achieve consensus. The following data were extracted: study characteristics (design, the first author, year of publication, country), source of strain (inpatients or outpatients), samples (type, number), intervention (previous processing, Bruker database version, procedures), and outcomes (correct identification rate, category of pathogens).
Quality assessment
The quality of the original studies was assessed independently by two investigators (BS and WW) by using the Quality Assessment of Diagnostic Accuracy Studies (QUADAS) guidelines (12): study design, self-established database, category of pathogens, reference methods, and blinded status.
Statistical analysis
Major effect size index was considered the correct identification ratio. Identification ratio was calculated as the number of isolates after correct identification by MALDI-TOF MS divided by the total number of isolates obtained with the reference method used in each reference. All analyses were performed with Stata Statistical/Data Analysis Software Package (version 11.0; Stata Corp LP, College Station USA). The correct identification ratio was calculated using the Freeman-Tukey transformation of inverse hyperbolic sine function (13). Heterogeneity between studies was evaluated quantitatively with the basis of I2, which is the heterogeneity measure of Cochran’s Q test. Statistical significance was defined as a P value of <0.1 and an I2 value of <50% for the qualitative Cochran’s Q and quantitative I2 measures. I2 values of 0%, 25%, 50%, and 75% indicate no, low, moderate, and high heterogeneity, respectively. Egger’s regression method was applied to check whether publication bias might have influenced the validity of the results. Subgroup analyses were performed in the random-effect model when high heterogeneity was presented. Heterogeneity and pooled correct identification ratios by subgroup analyses at the species level were determined according to source of strains, category of strains, urine flow cytometry system, and different agar plates for culture.
Results
Eligible studies
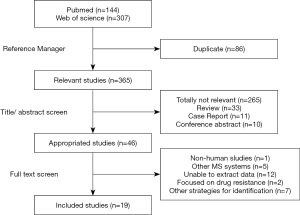
A total of 451 articles were obtained by searching the Web of Science and PubMed databases with the defined retrieval criteria. After duplicate removal using Thomson Reuters EndNote X9 and title/abstract review, 46 articles were retained for full-text scanning. A total of 265 references were excluded because they were totally not relevant to the study. In addition, 33 reviews, 11 case reports, and 10 conference abstracts were excluded. After the papers were screened, 1 study was excluded because organisms were not isolated from humans; 5 were excluded due to unrelated system used; 12 were rejected because of lack of detailed description of isolates; 2 were excluded due to their relevance to drug resistance; 7 were excluded due to other strategies employed for bacterial identification. As a result, 19 articles were included in the meta-analysis (Figure 1).
Quality of studies
The major characteristics of the enrolled eligible articles are listed in Table 1. Among the 19 articles, 12 were carried out in Europe (11 in Spain and 1 in France), 3 were performed in the USA, and 4 were conducted in Asia. All the studies reported on the identification performance of the Bruker’s BioTyper system allowing for greater standardization in evaluation (4-5,8-10,14-27). Among the 19 enrolled articles, 5 (26.31%) were prospective and 14 (73.69%) were retrospective. Only one article (23) stated that a blinded method was used, while the other studies did not specify the use of a blinded method in their investigation. Furthermore, one study (10) created a specific reference database by establishing the reference MALDI-TOF spectra of infected urine samples, while other studies used databases from instrument suppliers. Two articles reported on the algorithms for microbiological diagnosis of UTIs through screening by luminometry and Gram-staining, followed by identification using MALDI-TOF MS (14,24). One study developed a special method for pretreatment of urine samples with SDS to increase the method sensitivity (25). One study evaluated Bruker MS with urine preparation by NaOH-HCl neutralization allowing for rapid and cost-effective bacterial identification (17). One study developed diafiltration MALDI-TOF MS method to detect and identify pathogens directly from urine specimens (23). Briefly, all references used clinical isolates, and the total number of included samples was 4,579.
Full table
Overall meta-analysis
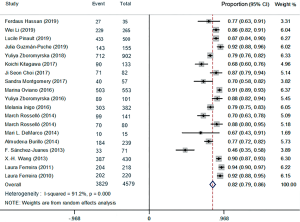
The forest plots of the random-effects model were used in meta-analysis to conclude the overall statistics at the species level. The overall correct identification ratio of MALDI-TOF MS for clinical pathogens in various studies ranged from 0.46 to 0.94. Significant heterogeneity was found at the species level (I2 =91.2%; P<0.0001). The random-effects pooled identification accuracy of MALDI-TOF MS was 0.82 with 95% CI of 0.79 to 0.86 (Figure 2).
The overall results for Gram-negative bacteria were reported in 15 references that included 3,150 samples. The forest plots of the random-effects model were used, and the correct identification ratio in individual studies ranged from 0.54 to 0.98. High heterogeneity was found at the species level (I2 =93.1%; P<0.0001). The estimated pooled correct identification rate was determined as 0.87 (95% CI: 0.83 to 0.91) by using the random-effects model. Fourteen studies that included 537 samples reported results for Gram-positive bacteria. The correct identification ratio for these pathogens ranged from 0.32 to 0.80 at the species level. Moderate heterogeneity was found in the meta-analysis (I2 =79.3%; P<0.0001), and the pooled correct identification ratio estimated using the random-effects model was 0.59 (95% CI: 0.49 to 0.68) (Figure 3).
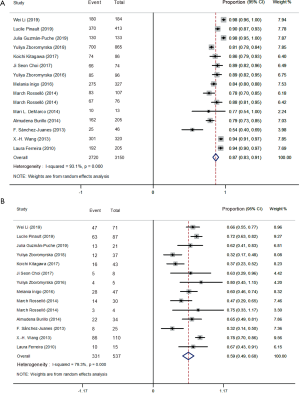
Meta-analysis of different pathogen species
The results for the correct identification of different pathogens are presented in Table 2. Gram-negative bacteria with the highest correct identification rates were of Morganella morganii, Enterobacter aerogenes, Serratia marcescens, Acinetobacter baumannii, and Citrobacter freundii. No heterogeneity was found in these species (I2 =0.0%, P>0.50). Escherichia coli and Klebsiella pneumoniae, which are the most important and common pathogens in UTIs, showed high identification proportions at 0.90 with 95% CI of 0.86 to 0.94 and 0.99 with a 95% CI of 0.98 to 1.00, respectively. The correct identification rates for other Gram-negative pathogens were 0.73 for Proteus mirabilis (95% CI: 0.49–0.98) and 0.96 for Pseudomonas aeruginosa (95% CI: 0.92–1.00) (Table 2).
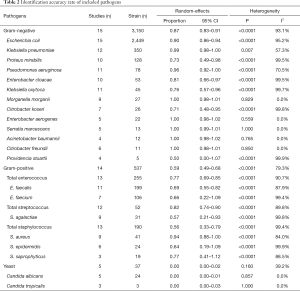
Full table
For Gram-positive bacteria, Enterococcus faecalis and Enterococcus faecium were the most commonly isolated microorganisms with moderate identification proportions at 0.69 (95% CI: 0.55 to 0.82) and 0.66 (95% CI: 0.22–1.09), respectively. The rates of correct identification of Staphylococcus aureus, Staphylococcus epidermidis, and Staphylococcus saprophyticus ranged from 0.64 to 0.94. The lowest direct bacterial identification proportion was that of Streptococcus agalactiae at 0.57 (95% CI: 0.21–0.93). Significant heterogeneity for each species of Gram-positive bacteria was found in the meta-analysis, with I2 values above 80% (P<0.0001).
In this meta-analysis, MALDI-TOF MS-based method showed poor performance in identification of yeasts. The estimated pooled correct identification ratio was 0.00.
Subgroup meta-analyses
Subgroup analyses for heterogeneity and correct identification rates were performed according to source of strains (inpatients only or inpatients plus outpatients), category of strains (Gram-negative bacteria, Gram-positive bacteria, and yeast), automated urine flow cytometry system (BacterioScan 216Dx, Sysmex UF-500i or Sysmex UF-1000i), and agar plates for culture (blood agar plate or cysteine lactose electrolyte deficient agar plate) (Table 3). The correct identification performance of the sub-analyses on Gram-negative isolates slightly exceeded the overall ratio (87% vs. 82%). Meanwhile, the rate of Gram-positive isolates was inferior to the overall ratio (59% vs. 82%). The correct identification performance for yeast at the species level was 0%. No significant difference was observed among different urine flow cytometry systems. The rates of the sub-analyses on isolates from inpatients only on blood agar plates were similar to their comparative group. Notably, the heterogeneity was not obviously decreased, except for BacterioScan 216Dx group and Sysmex UF-500i group in the subgroup meta-analyses.
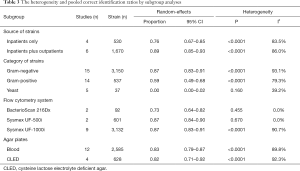
Full table
Assessment of publication bias
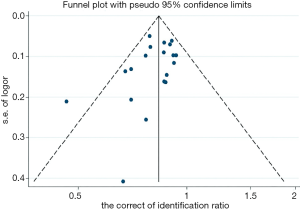
The quality of all studies included was moderate. Minimal publication bias was detected by Egger’s linear regression test of funnel plot asymmetry (t=−1.54 and P=0.143 for Egger’s) at the species level (Figure 4).
Discussion
MALDI-TOF MS, which is based on the identification of characteristic protein profiles for each microorganism, has made considerable achievements. As a newly developed technology in clinical microbiology laboratories, MALDI-TOF MS has been proven as a very rapid, accurate, and economical technique for bacterial identification compared with conventional methods. MALDI-TOF MS has undoubtedly changed the manner through which infectious diseases are diagnosed in clinical settings (28). The first application of MALDI-TOF MS dated back to 1975, but it was not entirely successful because bacteria were not identified at the species level (29). In the 1980s, with the advent of soft ionization techniques, bacterial identification using MALDI-TOF MS became possible. At present, MALDI-TOF MS is applied to new fields, such as biomolecular changes and protein profiling. The use of MALDI-TOF MS-based methods for direct bacteria identification from clinical specimens, such as urine and blood samples, has been developed. Conventional phenotypic and biochemical methods, molecular microbiological techniques, and MALDI-TOF MS are all important components of clinical laboratories. The unique advantage of MALDI-TOF MS over traditional approaches is the significant reduction in TAT. MALDI-TOF MS has significant impact on cost savings and public health when compared with molecular microbiological techniques that employ complicated procedures and have high cost.
The effectiveness of MALDI-TOF MS-based methods for direct identification of pathogens in clinical specimens, such as blood and sterile body fluids, has been reported (30,31). Previous studies indicated that urine is an optimal specimen for direct MALDI-TOF MS-based identification (4). The present systematic review and meta-analysis synthesized the available studies to evaluate the validity of MALDI-TOF MS for direct identification of bacteria from positive urine specimens. According to the selection criteria, 19 articles were included in the meta-analysis. We mainly focused on positive samples that had significant growth of microorganism by urine culture. The pooled results demonstrated that MALDI-TOF MS is a rapid and reliable method for direct identification of UTI pathogens from urine samples, especially in cases of Gram-negative bacterial infections. The overall correct identification ratio for clinical pathogens in various studies was 0.82 (95% CI: 0.79 to 0.86) at the species level.
MALDI-TOF MS showed highly accurate identification of Gram-negative bacteria at the species level and moderate accuracy for identification of Gram-positive bacteria, with 0.87 (95% CI: 0.83 to 0.91) versus 0.59 (95% CI: 0.49 to 0.68) bacteria identified reliably, respectively. The method exhibited superior correct identification ratio for Gram-negative pathogens than for Gram-positive pathogens. Several Gram-positive pathogens are difficult to identify possibly because of their thicker and highly anionic cell wall structure, which acts as permeability barrier, resulting in decreased protein extraction (32). This phenomenon might be explained by the unsuccessful removal of urine cells during sample processing (9).
Escherichia coli (2,449/4,579, 53.48%) was the most common pathogen in the 19 articles included in the meta-analysis, showing a high identification proportion at 0.90 with a 95% CI of 0.86 to 0.94. The results demonstrated that MALDI-TOF MS had strong capability for direct pathogen identification from the vast majority of urine samples of patients with UTI. Incorporating this technology in clinical laboratories could satisfy the demand of rapid diagnosis for specific patients, such as pediatric newborns or kidney transplant patients. The identification performances of MALDI-TOF MS for several pathogen species varied. For several relatively unusual isolates, the identification proportion ranged from 0.50 to 1.00. This observation may be due to the inherent difficulty of obtaining satisfactory spectra from unusual isolates and the insufficient number of spectra in commercial reference databases (33). Sub-analysis was performed according to categories of strains, sources of strains, flow cytometry systems, and agar plates. No significant difference was observed among different urine flow cytometry systems, different sources of strains, and different agar plates for culture. These results indicated that the accuracy of MALDI-TOF MS for direct bacterial identification from culture-positive urine specimens is not affected by the type of urine flow cytometry used for sample screening, the strains from inpatients or outpatients, and the selection of agar plates. However, the method showed poor performance in identification of yeasts. In general, clinical pathogenic yeasts are not easy to identify by MALDI-TOF MS in positive blood cultures as well as in positive urine cultures (34). In this article, only 5 studies (9,14,17,21,24) that included 37 samples presented the results of yeasts, and the pooled correct identification ratio was 0.00 with 95% CI of 0.00 to 0.02. On the one hand, the protein extraction protocols for yeasts are not standardized. On the other hand, compared with bacteria, yeasts exhibit radically different phenotypes and protein spectra depending on their growth conditions or the zone of the mycelium (35). Therefore, rapid identification of yeasts directly from urine samples remains a challenge for efficient diagnosis of UTI.
Previous studies reported that the main pathogens of UTI are Gram-negative bacilli, and the dominant pathogens are Escherichia coli and Klebsiella pneumoniae (27). These common isolates are easier to identify than complicated UTI pathogens. Nevertheless, a number of solutions have been proposed in literature. One study proposed an algorithm based on performing Gram staining followed by MALDI-TOF MS. The serial diagnostic algorithm improved the identification accuracy of UTI pathogens, especially when the causative microorganisms have resistance mechanisms (i.e., betalactamase-producing Enterobacteriaceae and yeasts). Urinary antibacterial activities have been increasingly reported. The presence of urinary antibacterial activities can affect the identification of bacteria in urine. Sánchez-Juanes et al. (25) improved the direct identification of bacteria in urine sample by pretreatment with SDS, a compound that might release microorganisms and increase the method sensitivity. Application of a protein extraction procedure to bacterial pellets has been reported to improve direct identification by MALDI-TOF MS. A microliter of 70% formic acid was added directly to the MALDI-TOF target sample to improve the spectral quality during bacterial identification.
In this meta-analysis, we excluded unpublished papers and gray literature to avoid possible bias in the application of selection criteria. We attempted to avoid this bias as much as possible by performing searches without publication status, geographical distribution, and language restrictions. Subgroup analyses of heterogeneity and correct identification rates were also performed according to source of strains, category of strains, and methods of identifying causative microorganisms.
In conclusion, this meta-analysis demonstrated that MALDI-TOF MS provides a reliable etiology basis for diagnosis of UTI, especially in cases of Gram-negative bacterial infections. In the future, the methodology should be optimized for identification of Gram-positive bacteria or yeasts that cause UTI.
Acknowledgments
Funding: This research was supported by the National Natural Science Foundation of China (81871734, 81471994), Jiangsu Provincial Natural Science Foundation (BK20151154), Jiangsu Provincial Medical Talent (ZDRCA2016053), Six talent peaks project of Jiangsu Province (WSN-135), Advanced health talent of six-one project of Jiangsu Province (LGY2016042).
Footnote
Reporting Checklist: The authors have completed the PRISMA reporting checklist. Available at: http://dx.doi.org/10.21037/atm-20-7310
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-7310). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Flores-Mireles AL, Walker JN, Caparon M, et al. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol 2015;13:269-84. [Crossref] [PubMed]
- Akoachere JF, Yvonne S, Akum NH, et al. Etiologic profile and antimicrobial susceptibility of community-acquired urinary tract infection in two Cameroonian towns. BMC Res Notes 2012;5:219. [Crossref] [PubMed]
- Mazzariol A, Bazaj A, Cornaglia G. Multi-drug-resistant Gram-negative bacteria causing urinary tract infections: a review. J Chemother 2017;29:2-9. [Crossref] [PubMed]
- Ferreira L, Sanchez-Juanes F, Gonzalez-Avila M, et al. Direct identification of urinary tract pathogens from urine samples by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 2010;48:2110-5. [Crossref] [PubMed]
- March Rosselló GA, Gutiérrez Rodríguez MP, Ortiz de Lejarazu Leonardo R, et al. New procedure for rapid identification of microorganisms causing urinary tract infection from urine samples by mass spectrometry (MALDI-TOF). Enferm Infecc Microbiol Clin 2015;33:89-94. [Crossref] [PubMed]
- Jolkkonen S, Paattiniemi EL, Karpanoja P, et al. Screening of urine samples by flow cytometry reduces the need for culture. J Clin Microbiol 2010;48:3117-21. [Crossref] [PubMed]
- Pieretti B, Brunati P, Pini B, et al. Diagnosis of bacteriuria and leukocyturia by automated flow cytometry compared with urine culture. J Clin Microbiol 2010;48:3990-6. [Crossref] [PubMed]
- Hassan F, Bushnell H, Taggart C, et al. Evaluation of BacterioScan 216Dx in Comparison to Urinalysis as a Screening Tool for Diagnosis of Urinary Tract Infections in Children. J Clin Microbiol 2019;57:00571. [Crossref] [PubMed]
- Li W, Sun EH, Wang Y, et al. Rapid Identification and Antimicrobial Susceptibility Testing for Urinary Tract Pathogens by Direct Analysis of Urine Samples Using a MALDI-TOF MS-Based Combined Protocol. Front Microbiol 2019;10:1182. [Crossref] [PubMed]
- Pinault L, Chabrière E, Raoult D, et al. Direct Identification of Pathogens in Urine by Use of a Specific Matrix-Assisted Laser Desorption Ionization-Time of Flight Spectrum Database. J Clin Microbiol 2019;57:01678. [Crossref] [PubMed]
- Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. BMJ 2009;339:b2700. [Crossref] [PubMed]
- Whiting P, Rutjes AW, Reitsma JB, et al. The development of QUADAS: a tool for the quality assessment of studies of diagnostic accuracy included in systematic reviews. BMC Med Res Methodol 2003;3:25. [Crossref] [PubMed]
- Ruiz-Aragón J, Ballestero-Téllez M, Gutiérrez-Gutiérrez B, et al. Direct bacterial identification from positive blood cultures using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry: A systematic review and meta-analysis. Enferm Infecc Microbiol Clin 2018;36:484-92. [Crossref] [PubMed]
- Guzmán-Puche J, Gracia-Ahufinger I, Causse M, et al. Combination of Coral UTI ScreenTM system, gram-stain and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for diagnosis of urinary tract infections directly from urine samples. J Chemother 2019;31:74-80. [Crossref] [PubMed]
- Zboromyrska Y, Bosch J, Aramburu J, et al. A multicentre study investigating parameters which influence direct bacterial identification from urine. PLoS One 2018;13:e0207822 [Crossref] [PubMed]
- Kitagawa K, Shigemura K, Onuma KI, et al. Improved bacterial identification directly from urine samples with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J Clin Lab Anal 2018;32:e22301 [Crossref] [PubMed]
- Choi JS, Kim J, Kim HS, et al. NaOH-HCl neutralized urine preparation for direct testing of uropathogens by Bruker MS. J Clin Lab Anal 2018;32:e22280 [Crossref] [PubMed]
- Montgomery S, Roman K, Ngyuen L, et al. Prospective Evaluation of Light Scatter Technology Paired with Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry for Rapid Diagnosis of Urinary Tract Infections. J Clin Microbiol 2017;55:1802-11. [Crossref] [PubMed]
- Oviaño M, Ramirez CL, Barbeyto LP, et al. Rapid direct detection of carbapenemase-producing Enterobacteriaceae in clinical urine samples by MALDI-TOF MS analysis. J Antimicrob Chemother 2017;72:1350-4. [Crossref] [PubMed]
- Zboromyrska Y, Rubio E, Alejo I, et al. Development of a new protocol for rapid bacterial identification and susceptibility testing directly from urine samples. Clin Microbiol Infect 2016;22:561.e1-561.e6. [Crossref] [PubMed]
- Íñigo M, Coello A, Fernández-Rivas G, et al. Direct Identification of Urinary Tract Pathogens from Urine Samples, Combining Urine Screening Methods and Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry. J Clin Microbiol 2016;54:988-93. [Crossref] [PubMed]
- March Rosselló GA, Gutiérrez Rodríguez MP, de Lejarazu Leonardo RO, et al. Procedure for microbial identification based on Matrix-Assisted Laser Desorption/Ionization-Time of Flight Mass Spectrometry from screening-positive urine samples. Apmis 2014;122:790-5. [Crossref] [PubMed]
- Demarco ML, Burnham CA. Diafiltration MALDI-TOF mass spectrometry method for culture-independent detection and identification of pathogens directly from urine specimens. Am J Clin Pathol 2014;141:204-12. [Crossref] [PubMed]
- Burillo A, Rodríguez-Sánchez B, Ramiro A, et al. Gram-stain plus MALDI-TOF MS (Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry) for a rapid diagnosis of urinary tract infection. PLoS One 2014;9:e86915 [Crossref] [PubMed]
- Sánchez-Juanes F, Siller Ruiz M, Moreno Obregón F, et al. Pretreatment of urine samples with SDS improves direct identification of urinary tract pathogens with matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 2014;52:335-8. [Crossref] [PubMed]
- Wang XH, Zhang G, Fan YY, et al. Direct identification of bacteria causing urinary tract infections by combining matrix-assisted laser desorption ionization-time of flight mass spectrometry with UF-1000i urine flow cytometry. J Microbiol Methods 2013;92:231-5. [Crossref] [PubMed]
- Ferreira L, Sanchez-Juanes F, Munoz-Bellido JL, et al. Rapid method for direct identification of bacteria in urine and blood culture samples by matrix-assisted laser desorption ionization time-of-flight mass spectrometry: intact cell vs. extraction method. Clinical Microbiology and Infection 2011;17:1007-12. [Crossref] [PubMed]
- Zhou C, Hu B, Zhang X, et al. The value of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in identifying clinically relevant bacteria: a comparison with automated microbiology system. J Thorac Dis 2014;6:545-52. [PubMed]
- Ko KH, Kwon CI, Park SH, et al. Application of Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Imaging Mass Spectrometry (MALDI-TOF IMS) for Premalignant Gastrointestinal Lesions. Clin Endosc 2013;46:611-9. [Crossref] [PubMed]
- Tian Y, Zheng B, Wang B, et al. Rapid identification and multiple susceptibility testing of pathogens from positive-culture sterile body fluids by a combined MALDI-TOF mass spectrometry and vitek susceptibility system. Front Microbiol 2016;7:523. [Crossref] [PubMed]
- Ferreira L, Sánchez-Juanes F, Porras-Guerra I, et al. Microorganisms direct identification from blood culture by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Clin Microbiol Infect 2011;17:546-51. [Crossref] [PubMed]
- Kline KA, Lewis AL. Gram-positive uropathogens polymicrobial urinary tract infection, and the emerging microbiota of the urinary tract. Microbiol Spectr 2016;4:e00122012 [Crossref] [PubMed]
- Ling H, Yuan Z, Shen J, et al. Accuracy of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of clinical pathogenic fungi: a meta-analysis. J Clin Microbiol 2014;52:2573-82. [Crossref] [PubMed]
- Rodríguez-Sánchez B, Sánchez-Carrillo C, Ruiz A, et al. Direct identification of pathogens from positive blood cultures using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. Clin Microbiol Infect 2014;20:O421-7. [Crossref] [PubMed]
- Steensels D, Verhaegen J, Lagrou K. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for the identification of bacteria and yeasts in a clinical microbiological laboratory: a review. Acta Clin Belg 2011;66:267-73. [PubMed]