
Potential functional variants of KIAA genes are associated with breast cancer risk in a case control study
Introduction
Breast cancer, one of the commonest malignancies, is responsible for the highest cancer-related mortality rate in women. In 2018, an estimated 2,088,849 new cases and 626,679 deaths occurred worldwide (1). Breast cancer development involves multiple stages and is influenced by genetics and the environment (2-4). The most prevalent risk factors are those linked to menstruation (menarche at a young age, older age at menopause), reproduction (nulliparity, first birth at a late age, and fewer offspring), artificial hormones (oral contraceptives and hormone-replacement therapies) and alcohol intake (2). Genome-wide association studies (GWASs) are known to be powerful tools for dissecting the genetic architecture of complex diseases. To date, GWASs have identified over 150 loci for breast cancer susceptibility in several populations (3,4). Despite this, the information provided by GWAS remains insufficient. The heritability of breast cancer is estimated to be responsible for only 18% of the familial relative risk of breast cancer (3). Moreover, most of the breast cancer susceptibility SNPs that have been identified in breast cancer GWAS studies are located in intergenic or intronic regions of the genome, which may not generally impact the protein-coding regions as expectations (5,6).
KIAA genes were identified by the Kazusa cDNA-sequencing project. Sequences of human large cDNAs longer than 4 kb which direct synthesis of large proteins (>50 kDa) were characterized and deposited in the HUGE database (Human Unidentified Gene-Encoded large protein database) (7,8). Because KIAA genes were screened and identified mainly based upon the length of the mRNA (>4 kb) that encode large proteins (>1,000 amino acid residues), KIAA proteins possess many kinds of biological functions and join in important life activities. Some KIAA proteins are thought to play critical roles in multiple biological processes, such as DNA damage, centriole formation, cell migration and invasion (9-11). Recently, the potential importance of KIAA genes in cancer is beginning to be recognized and studied, which has attracted considerable attention as a target for various biological events in tumorigenesis. A growing number of evidences have shown that abnormal expression of KIAA genes were associated with the development and prognosis of various cancers, such as gastric cancer, esophageal squamous-cell carcinomas, colorectal cancer, lung cancer and breast cancer (12-16). Abnormal increase or loss of KIAA genes expression may contribute to tumor formation and development. Kang et al. observed that KIAA1324 not only inhibited the invasiveness, growth, and tumorigenicity of gastric cancer cells but also blocked oncogenic GRP78 activities to promote apoptosis. Low KIAA1324 expression was related to poor prognosis in patients with gastric cancer (12). KIAA1377 has been found to be significantly amplified in esophageal squamous cell carcinoma with lymph node metastasis (13). KIAA1199 has been identified as an oncogene in many cancers including breast cancer and associated with tumor invasion depth, TNM stage, and poor prognosis (14-16).
In recent year, polymorphisms in KIAA genes have drawn considerable attention in human complex diseases, such as cancers, diabetes, autoimmune diseases and cardiac disease, and extensive efforts have been made to elucidate the mechanisms responsible for their aberrant expressions (17-22). However, up to now, there are few records of the association of single nucleotide polymorphisms (SNPs) in KIAA genes and breast cancer risk. In this study, we hypothesized that there is an association between polymorphisms of crucial KIAA genes and the incidence of breast cancer in the Chinese population. To test this hypothesis, we identified a number of differentially expressed KIAA genes potentially associated with breast cancer from The Cancer Genome Atlas (TCGA) research database, and adopted a case-control design to investigate the occurrence of the SNPs among Chinese females (1,032 cases and 1,063 controls). We expected to accumulate useful data to reveal the correlation between genetic polymorphisms of key KIAA genes and breast cancer risk.
We present the following article in accordance with the MDAR checklist and STROBE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-6108).
Methods
Study subjects
This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and received approval from the Institutional Review Board of Nanjing Medical University (FWA00001501). A total of 1,032 cases with clinically confirmed breast cancer and 1,063 cancer-free controls were included in the association analysis. All study participants were Chinese Han females, genetically unrelated to each other. The study design has been described in detail in a previous publication (23). Briefly, we included female patients who were histologically diagnosed with breast cancer at the First Affiliated Hospital of Nanjing Medical University, Cancer Hospital of Jiangsu Province, and Gulou Hospital, between 2004 and 2010. Controls were recruited from a noninfectious diseases’ community screening program between 2004 and 2006 in Jiangsu Province. We obtained informed consent from each participant before recruitment. Each participant was interviewed using a structured questionnaire to collect information on age, menstrual and reproductive history, and environmental exposure history. Clinicopathologic data comprising estrogen-receptor (ER) and progesterone-receptor (RP) statuses of patients were collected from their medical records. The demographic distribution in terms of ages between cases and controls was similar (±5 years).
SNP selection and genotyping
A total of 96 KIAA genes were selected from the Gencode databases (Table S1). Then we detected 65 of 96 KIAA genes with differential expression levels in normal samples compared with tumor samples using the breast cancer TCGA database (Student’s t-test, P<0.05) (Table S2).
A detailed workflow chart of the strategy has been stated as Figure 1. We screened the SNPs in 65 candidate KIAA genes from the 1000 Genomes Project according to the following criteria: (I) Population data for the Chinese Han population of Beijing in the 1000 Genomes Project was used to screen SNPs within the candidate KIAA gene regions and 2 kb upstream. A total of 12,323 SNPs remained after the following two quality control criteria were applied: call rate >95% and minor allele frequency ≥0.05. (II) We excluded SNPs that were not within a promoter, exon, or 3ʹ-/5ʹ-untranslated region. (III) HaploView 4.2 software was employed to extract SNPs that had linkage disequilibrium in the Chinese Han population (r2 ≥0.8). HaploReg (http://archive.broadinstitute.org/mammals/haploreg/haploreg.php) and RegulomeDB (http://regulome.stanford.edu/index) were used to evaluate SNPs for regulatory elements and potential functions.
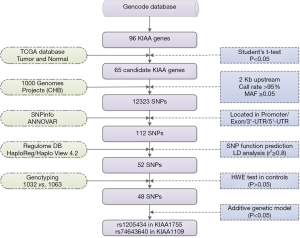
In total, 52 SNPs located in KIAA gene regions were selected for further genotyping. We performed genotyping using the Illumina HumanExome Beadchip array (Illumina Inc.) in 1,064 breast cancer cases and 1,125 cancer-free controls. Genotyping was performed without knowledge of the individual’s case or control status, and approximately equal numbers of case and control samples were tested during each assay, with two blank controls. Genotype calling was performed using Illumina’s GenTrain clustering algorithm (version 1.0) in GenomeStudio (V2011.1). Thirty-two cases and sixty-two control subjects were excluded by filtering unqualified genetic variants and genetic relatedness. The study design has been described in detail in a previous publication (23). Finally, 1,032 breast cancer cases and 1,063 cancer-free controls were genotyped for SNPs by Illumina HumanExome Beadchip array. Meanwhile, Hardy-Weinberg equilibrium was used to filter SNPs with P values less than 0.05 in control groups and 4 SNPs were excluded. Lastly, 48 SNPs located in the 28 KIAA genes were left for further analysis.
Statistical analyses
For each SNP, a χ2 test using one degree of freedom was employed to examine Hardy-Weinberg equilibrium (HWE) in controls. We used a χ2 test and student’s t-test to calculate the differences between the chosen categorical and continuous variables, respectively, the genotypic frequency of each of the 52 SNPs in the controls and cases, and differences in the distribution of demographic characteristics. Confidence intervals (CIs, 95%) and odds ratios (ORs) were established using logistical regression for the associations between a given SNP and breast cancer risk adjusted for menopausal status, age at menarche, as well as current age. Heterogeneity of associations between subgroups was assessed by the χ2-based Q test. Interaction analysis was performed by adding an interaction item in logistic regression model. All statistical analyzes were performed with R software (version 3.2.3) for Statistical Computing. Statistical significance was indicated by a two-sided P value of <0.05.
Bioinformatics analysis
SNPs influencing the protein stability of KIAA genes were analyzed using I-Mutant (http://folding.biofold.org/i-mutant/i-mutant2.0.html) and MUpro (http://mupro.proteomics.ics.uci.edu/).
We assessed the eQTL of the candidate SNPs using the Gene-Tissue Expression (GTEx) project (http://www.gtexportal.org/). Fully processed, filtered, and normalized gene expression matrices (in BED format), as well as covariates, were download from the GTEx website. Then, genotype information from the GTEx Analysis V8 (dbGaP Accession phs000424.v8.p2) was applied from the dbGap database for eQTL analysis.
Online Kaplan-Meier plotter
The Kaplan-Meier plotter (http://kmplot.com/analysis/) was used to investigate how KIAA-gene expression affected the survival of breast cancer patients. We obtained the expression data for the candidate genes and overall patient survival data from the public databases. We analyzed the gene’s prognostic values by allocating the patient samples into two groups, which were then compared using Kaplan-Meier curves. The hazard ratios (HRs), the corresponding 95% confidence intervals (CIs), and log-rank P values were generated.
Results
Characteristics of the study population
The demographic characteristics of 1,032 breast cancer cases and 1,063 cancer-free controls are described in Table S3. There were no significant differences in the distribution of ages between the cancer and control groups (P=0.078). The analysis showed that women with breast cancer went through menarche at a significantly younger age than the control group (P<0.0001), and birthed their first live child at a significantly elder age than the control group (P<0.0001).
Association between candidate SNPs in KIAA genes regions and breast cancer risk
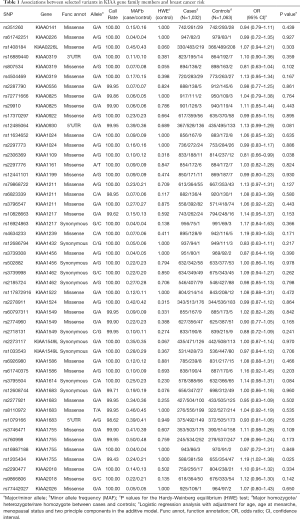
This study primarily aimed to investigate the possible associations between 48 predominant SNPs (r2<0.8) in 28 candidate KIAA genes and the risk of breast cancer. Through the additive model, we found two SNPs, rs1205434 and rs2306369, that were significantly related to breast cancer risk in our population (P<0.05) (Table 1). According to the additive model, there was a significant association between rs1205434 in the KIAA1755 coding region and increased breast cancer incidence (rs1205434 adjusted OR =1.19, 95% CI: 1.02–1.38, P=0.025). In contrast, the model showed there was a significant association between rs2306369 in the KIAA1109 coding region and a decrease in breast cancer incidence (rs2306369 adjusted OR =0.81, 95% CI: 0.66–0.99, P=0.038). All statistical analyzes were adjusted for possible influential confounders, i.e., age, age at menarche, and menopausal status.
Full table
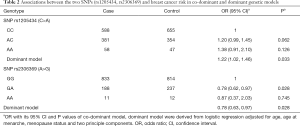
Additionally, under the dominant and codominant models, correlations between the risk of breast cancer and the two SNPs (rs1205434 and rs2306369) were also calculated. Table 2 lists further details. The results showed that the SNP rs1205434 was significantly correlated with increased breast cancer individual risk under the dominant model (OR =1.22, 95% CI: 1.02–1.46, P=0.033); whereas the SNP rs2306369 was significantly correlated with decreased breast cancer individual risk under both the codominant and dominant models (codominant: OR =0.78, 95% CI: 0.62–0.97, P=0.028; dominant: OR =0.78, 95% CI: 0.63–0.97, P=0.028).
Full table
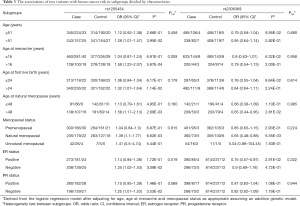
We also performed stratification analyses of rs1205434 and rs2306369 based on the age group, age at menarche, age at first live birth, age at natural menopause, and menopause status. As shown in Table 3, the SNPs rs1205434 and rs2306369 exhibited different effects among women with later natural menopause and at different menopause stages (P<0.05). Additionally, rs1205434 showed a significant difference for age at menarche strata (heterogeneity test P=0.009). Further multiplicative interaction analysis indicated that there may exist an interaction effect between rs1205434 and menarche age (Table 4). Women who both carry CA/AA genotype of rs1205434 and menarche over 16 years would prone to develop breast cancer (OR =1.09, 95% CI: 1.01–1.17, P=0.036).
Full table
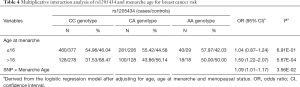
Full table
Potential functional impact of KIAA genes variants
In silico annotation showed that the SNPs for KIAA1109 (rs2306369) and KIAA1755 (rs1205434) were non-synonymous. The two SNPs were identified as those that decrease protein stability using I-Mutant and Mupro online bioinformatics analyzes (Table S4).
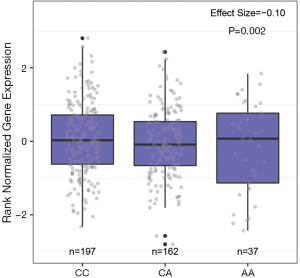
We examined if the two SNPs were eQTLs in the GTEx database. As shown in Figure 2, the SNP rs1205434 was in the eQTL for KIAA1755 (P=2.2×10−3) in normal breast tissue. While, according to eQTL analysis, rs2306369 was not significantly associated with the expression of its host gene KIAA1109 and nearby genes (±1MB).
KIAA gene expression and breast cancer survival
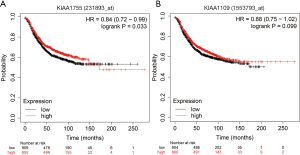
We assessed the relationships between KIAA gene expression and the prognosis of patients with breast cancer using the Kaplan-Meier plotter (http://kmplot.com/analysis/), which analyzes a combination of follow-up information and gene expression data (24). Patients were allocated to high- and low-expression groups based on KIAA gene expression mean values. The log-rank of the Kaplan Meier curve showed that the outcomes of patients with breast cancer who had high KIAA1755 expression were significantly better than those who had low KIAA1755 expression (HR =0.84, 95% CI: 0.72–0.99, P=0.033), while KIAA1109 expression showed no significant differences between the two groups (Figure 3).
Discussion
In this case-control study, we investigated 48 SNPs of KIAA genes linked to incidences of breast cancer risk. Two coding missense variant SNPs (rs2306369 and rs1205434) in the coding regions of KIAA1109 and KIAA1755 respectively were significantly associated with susceptibility to breast cancer in a Han Chinese population. Moreover, we found that rs1205434 genotype correlated with KIAA1755 expression by eQTL analysis. Additionally, elevated KIAA1755 expression may be associated with better breast cancer patient outcomes through a Kaplan-Meier plotter-based analysis.
The SNP rs2306369 is a coding variant (encoding p. Thr4440Ala) of the KIAA1109 gene. KIAA1109, which has been named Tweek, is conserved in many eukaryotes, from nematodes to vertebrates, although little is known of its function. KIAA1109 deficiency was found to be lethal in mice, who died prior to weaning; one possible function of the protein is in synaptic vesicle recycling, as has been found in Drosophila (25). Gueneau et al. reported finding KIAA1109 variants in individuals with severe brain-development disorders and arthrogryposis, and the report discusses the possible involvement of KIAA1109 in cell-cycle control mechanisms, especially those of the central nervous system (26). Additionally, mutations in KIAA1109 have been linked to survival in patients with endometrial cancer and esophageal squamous-cell carcinomas. Furthermore, bioinformatics studies showed KIAA1109 was involved in regulating NIK/NF-kappaB signaling (27,28).
KIAA1109 is encoded on chromosomal region 4q27; this region and the KIAA1109-interleukin 2 (IL2)-IL21 block, in particular, was identified as a possible locus of risk in the development of a number of common inflammatory disorders, such as type I diabetes, ulcerative colitis, systemic lupus erythematosus, celiac disease, Chron’s disease, psoriasis, and rheumatoid arthritis, via GWASs and candidate gene studies (19-21,29-32). In addition, the SNP rs13151961 in the intron of KIAA1109 gene has been implicated in male susceptibility to prostate cancer in those with a family history of the disease (17). The results reported herein reveal a significant association between the coding variant SNP rs2306369 and a decreased risk of breast cancer. Moreover, we found that the risk effects of rs2306369 variant genotypes were statistically significant in some subgroups, such as in women with later menopause, natural menopause, and ER-positive individuals. MUpro and I-Mutant software predicted that the protein structure stability of KIAA1109 would be decreased by the missense variant rs2306369. Detailed functional assays are required to validate these findings.
The missense variant rs1205434 is located within the KIAA1755 gene and encodes p.Lys339Asn. KIAA1755 is predicted to encode an uncharacterized protein and little known its function. Antony et al. found that KIAA1755 is a tumor-specific antigen found in the ascitic fluid of women with ovarian cancer (33). In this study, we investigated KIAA1755 expression and its effect on breast cancer prognosis through the online Kaplan-Meier plotter. The results showed that patients with high expression of KIAA1755 have relatively improved survival rates. Furthermore, levels of KIAA1755 were lower in tumors compared with in adjacent normal tissue, according to the TCGA breast cancer database. The results suggest KIAA1755 may play a tumor-suppressor role in breast cancer.
In the current literature, some reports state that KIAA1755 genetic variants may be associated with heart rate and hypertension. For example, the SNP rs6123471 in the 3ʹ-UTR of KIAA1755 was associated, at a genome-wide significance level, with heart rate variability in individuals of European ancestry (22). Exome chip meta-analyses identified a relationship between a low-frequency and non-synonymous single-nucleotide variant, rs41282820, at locus KIAA1755, and heart rate (34). Loss-of-function mutations in the KIAA1755 DNA sequence elevated eicosapentaenoate levels. Eicosapentaenoate, an essential fatty acid, has been found to increase essential hypertension in patients (35). The results of the present study revealed a significant association between non-synonymous SNP rs1205434 and an increase in the incidence of breast cancer. Moreover, we found that the risk effects of rs1205434 variant genotypes were statistically significant in some subgroups, such as older women and those with later menarche, later at first live birth, later menopause, etc. Additionally, multiplicative interaction analysis indicated that there may exist an interaction effect between rs1205434 and menarche age. According to the online bioinformatics analysis, rs1205434 (Lys339Asn) showed benign amino acid changes, and it probably damages KIAA1755 stability. Furthermore, compared with the C allele of rs1205434, the A allele showed a significant association with decreased KIAA1755 mRNA levels in mammary tissues with eQTL analysis. Together these results thus suggested that the rs1205434 may be a potential predictive biomarker for breast cancer occurrence, although further functional assays will be needed to confirm this result.
In conclusion, we studied the relationships between 48 SNPs of KIAA genes and the incidence of breast cancer in a population of Chinese Han women. Our study showed that the representative genetic variants rs2306369 and rs1205434 in the respective coding regions of KIAA1109 and KIAA1755 are candidate SNPs for use as markers for breast cancer susceptibility in this population. However, a few limitations of this study should be addressed. Firstly, we conducted one stage case control study with 1032 breast cancer cases and 1063 healthy controls. Studies with large sample size and more diverse different populations are needed to validate the results. Secondly, although we found bioinformatics evidence, the biological effect of the two SNPs (rs2306369 and rs1205434) on KIAA1109 and KIAA1755 expression and function were not investigated in this study. Further studies, incorporating functional evaluations, of diverse ethnic populations are warranted to confirm these findings.
Acknowledgments
We thank the study participants and research staff for their contributions and commitment to this study.
Funding: This study was supported by grants from the National Natural Science Foundation of China (81521004), the Priority Academic Program for the Development of Jiangsu Higher Education Institutions (Public Health and Preventive Medicine), and the Top-notch Academic Programs Project of Jiangsu Higher Education Institutions (PPZY2015A067).
Footnote
Reporting Checklist: The authors have completed the MDAR checklist and STROBE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-6108
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-6108
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-6108). The authors declare that they have no conflict of interest regarding the publication of this paper.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Review Board of Nanjing Medical University (FWA00001501) and informed consent was taken from each participant before recruitment.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Baselga J, Norton L. Focus on breast cancer. Cancer Cell 2002;1:319-22. [Crossref] [PubMed]
- Milne RL, Kuchenbaecker KB, Michailidou K, et al. Identification of ten variants associated with risk of estrogen-receptor-negative breast cancer. Nat Genet 2017;49:1767-78. [Crossref] [PubMed]
- Michailidou K, Lindström S, Dennis J, et al. Association analysis identifies 65 new breast cancer risk loci. Nature 2017;551:92-4. [Crossref] [PubMed]
- Fachal L, Aschard H, Beesley J, et al. Fine-mapping of 150 breast cancer risk regions identifies 191 likely target genes. Nat Genet 2020;52:56-73. [Crossref] [PubMed]
- Ferreira MA, Gamazon ER, Al-Ejeh F, et al. Genome-wide association and transcriptome studies identify target genes and risk loci for breast cancer. Nat Commun 2019;10:1741. [Crossref] [PubMed]
- Suyama M, Nagase T, Ohara O. HUGE: a database for human large proteins identified by Kazusa cDNA sequencing project. Nucleic Acids Res 1999;27:338-9. [Crossref] [PubMed]
- Kikuno R, Nagase T, Nakayama M, et al. HUGE: a database for human KIAA proteins, a 2004 update integrating HUGEppi and ROUGE. Nucleic Acids Res 2004;32:D502-4. [Crossref] [PubMed]
- Polato F, Rusconi P, Zangrossi S, et al. DRAGO (KIAA0247), a New DNA Damage–Responsive, p53-Inducible Gene That Cooperates With p53 as Oncosupprossor. J Natl Cancer Inst 2014;106:dju053 [Crossref] [PubMed]
- Evensen NA, Kuscu C, Nguyen HL, et al. Unraveling the Role of KIAA1199, a Novel Endoplasmic Reticulum Protein, in Cancer Cell Migration. J Natl Cancer Inst 2013;105:1402-16. [Crossref] [PubMed]
- Tsuchiya Y, Yoshiba S, Gupta A, et al. Cep295 is a conserved scaffold protein required for generation of a bona fide mother centriole. Nat Commun 2016;7:12567. [Crossref] [PubMed]
- Kang JM, Park S, Kim SJ, et al. KIAA1324 Suppresses Gastric Cancer Progression by Inhibiting the Oncoprotein GRP78. Cancer Res 2015;75:3087-97. [Crossref] [PubMed]
- Zheng ST, Yang CC, Liu Q, et al. KIAA1377 is associated with lymph node metastasis in esophageal squamous cell carcinoma. Oncol Lett 2016;12:5223-8. [Crossref] [PubMed]
- Xu J, Liu Y, Wang X, et al. Association between KIAA1199 overexpression and tumor invasion, TNM stage, and poor prognosis in colorectal cancer. Int J Clin Exp Pathol 2015;8:2909-18. [PubMed]
- Jami MS, Hou J, Liu M, et al. Functional proteomic analysis reveals the involvement of KIAA1199 in breast cancer growth, motility and invasiveness. BMC Cancer 2014;14:194. [Crossref] [PubMed]
- Deng F, Lei J, Zhang X, et al. Overexpression of KIAA1199: An independent prognostic marker in nonsmall cell lung cancer. J Cancer Res Ther 2017;13:664-8. [Crossref] [PubMed]
- Tindall EA, Hoang HN, Southey MC, et al. The 4q27 locus and prostate cancer risk. BMC Cancer 2010;10:69. [Crossref] [PubMed]
- Cheng I, Plummer SJ, Neslund-Dudas C, et al. Prostate cancer susceptibility variants confer increased risk of disease progression. Cancer Epidemiol Biomarkers Prev. 2010;19:2124-32. [Crossref] [PubMed]
- Zhernakova A, Alizadeh BZ, Bevova M, et al. Novel association in chromosome 4q27 region with rheumatoid arthritis and confirmation of type 1 diabetes point to a general risk locus for autoimmune diseases. Am J Hum Genet 2007;81:1284-8. [Crossref] [PubMed]
- Sawalha AH, Kaufman KM, Kelly JA, et al. Genetic association of interleukin‐21 polymorphisms with systemic lupus erythematosus. Ann Rheum Dis 2008;67:458-61. [Crossref] [PubMed]
- Liu Y, Helms C, Liao W, et al. A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci. PLoS Genet 2008;4:e1000041 [Crossref] [PubMed]
- Nolte IM, Munoz ML, Tragante V, et al. Genetic loci associated with heart rate variability and their effects on cardiac disease risk. Nat Commun 2017;8:15805. [Crossref] [PubMed]
- Zhou W, Jiang Y, Zhu M, et al. Low-frequency nonsynonymous variants in FKBPL and ARPC1B genes are associated with breast cancer risk in Chinese women. Mol Carcinog 2017;56:774-80. [Crossref] [PubMed]
- Lánczky A, Nagy A, Bottai G, et al. For a general citation of the KM-plotter, please use: miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2,178 breast cancer patients. Breast Cancer Res Treat 2016;160:439-46. [PubMed]
- Verstreken P, Ohyama T, Haueter C, et al. Tweek, an evolutionarily conserved protein, is required for synaptic vesicle recycling. Neuron 2009;63:203-15. [Crossref] [PubMed]
- Gueneau L, Fish RJ, Shamseldin HE, et al. KIAA1109 Variants Are Associated with a Severe Disorder of Brain Development and Arthrogryposis. Am J Hum Genet 2018;102:116-32. [Crossref] [PubMed]
- Qing T, Zhu S, Suo C, et al. Somatic mutations in ZFHX4 gene are associated with poor overall survival of Chinese esophageal squamous cell carcinoma patients. Sci Rep 2017;7:4951. [Crossref] [PubMed]
- Qiao Z, Jiang Y, Wang L, et al. Mutations in KIAA1109, CACNA1C, BSN, AKAP13, CELSR2, and HELZ2 Are Associated With the Prognosis in Endometrial Cancer. Front Genet 2019;10:909. [Crossref] [PubMed]
- van Heel DA, Franke L, Hunt KA, et al. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat Genet 2007;39:827-9. [Crossref] [PubMed]
- Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661-78. [Crossref] [PubMed]
- Festen EA, Goyette P, Scott R, et al. Genetic variants in the region harbouring IL2/IL21 associated with ulcerative colitis. Gut 2009;58:799-804. [Crossref] [PubMed]
- Hollis-Moffatt JE, Gearry RB, Barclay ML, et al. Consolidation of evidence for association of the KIAA1109-TENR-IL2-IL21 rs6822844 variant with Crohn's disease. Am J Gastroenterol 2010;105:1204-5. [Crossref] [PubMed]
- Antony F, Deantonio C, Cotella D, et al. High-throughput assessment of the antibody profile in ovarian cancer ascitic fluids. Oncoimmunology 2019;8:e1614856 [Crossref] [PubMed]
- van den Berg ME, Warren HR, Cabrera CP, et al. Discovery of novel heart rate-associated loci using the Exome Chip. Hum Mol Genet 2017;26:2346-63. [Crossref] [PubMed]
- Yazdani A, Yazdani A, Elsea SH, et al. Genome analysis and pleiotropy assessment using causal networks with loss of function mutation and metabolomics. BMC Genomics 2019;20:395. [Crossref] [PubMed]


