
Nucleotide polymorphism in ARDS outcome: a whole exome sequencing association study
Introduction
Acute respiratory distress syndrome (ARDS) is characterized by increased pulmonary vascular permeability and reduced aerated lung tissue (1). With an extremely high hospital mortality among 35–46% (2), current therapeutic strategies to increase ARDS survival are still limited. Advances in etiology and pathology of ARDS are urging. Clinical factors and protocolized therapeutic strategy poorly explain ARDS outcome, the role of genetic locus in the pathogenesis of ARDS is increasingly recognized (3-6).
Numerous genetic variants were identified associated with ARDS outcome. Morrell (7) found that genetic variation in MAP3K1 associated with ventilator-free days in ARDS, while Wei (8) showed that the missense genetic variant in LRRC16A/CARMIL1 improved survival by attenuating platelet count decline in ARDS patients. However, as a heterogeneous disease with multiple and interactive pathogenic processes, the effect of genetics contributing differently (7-11), meanwhile, the racial and ethnic differences in mortality also exist (12,13). More than five categories of genes were found to associate with ARDS outcome: genes influencing immune regulation, genes influencing endothelial barrier function, genes influencing respiratory epithelial function, genes influencing coagulation, genes influencing injury and oxidative stress and so forth. Then a few genetic risk factors have been discovered by large-scale genotyping approaches, from in vivo or in vitro models of lung injury, which highlight the importance of identifying genetic biomarkers of ARDS outcome to further improve stratification. The mutational landscape and variability at single nucleotide polymorphisms (SNP) with ARDS outcome in Chinese is unknown, not to mention their associations. By whole-exome sequencing association study, our goal was to explore the associations between genetic variants and ARDS outcome.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-5728).
Methods
Setting
This was an investigator-initiated, single-center, prospective observational trial that was conducted in the intensive care unit of a tertiary care teaching hospital. The study protocol was approved by the Ethics Committee (Approval Number: 2015ZDSYLL014.0) of Zhongda Hospital, School of Medicine, Southeast University, and written informed consent was obtained from each patient or their next of kin. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Patients
Adult ARDS (according to Berlin definition) patients were enrolled in the trial. The diagnostic criteria included: (I) within one week of a known clinical insult or new or worsening respiratory symptoms; (II) chest imaging showing that bilateral opacities-not fully explained by effusions, lobar/lung collapse, or nodules; (III) respiratory failure not fully explained by cardiac failure or fluid overload; and (IV) arterial partial pressure of oxygen/fraction of inspiration oxygen (PaO2/FiO2 ratio, P/F ratio) less than or equal to 300 mmHg.
Data collection
Baseline-recorded data included demographic characteristics, comorbidities, and the origin and etiology of ARDS were collected by trained investigators. Severity of illness was assessed with the Acute Physiology and Chronic Health Evaluation (APACHE) II score within 24 hours on enrollment. Sequential Organ Failure Assessment (SOFA) and Murray lung injury score within 24 hours on enrollment were also calculated.
Predisposing conditions of ARDS were collected, and subphenotypes of ARDS were determined. Severe ARDS group and non-severe group were divided according to the severity of lung injury (Berlin definition). Patients with risk factors of pneumonia (pulmonary sepsis), pulmonary contusion, inhalation and drowning were categorized as having pulmonary ARDS, whereas patients with risk factors of non-pulmonary sepsis or pancreatitis were categorized as having extrapulmonary ARDS. Patients with sepsis on enrollment after enrolled were recorded as ARDS with sepsis. Patients with shock on enrollment were recorded as ARDS with shock. Sepsis was defined by Sepsis 3.0.
Peripheral blood samples were drawn. Prognosis was recorded as the survivors and non-survivors in ICU.
Study methods
Whole-exome sequencing was performed by the sequencing platform Illumina, the data were compared with reference genome UCSC hg19. Firstly, genomic DNA was isolated from the peripheral blood samples taken from individuals by following the manufacturer’s standard procedure using QIAamp DNA Blood kits (Qiagen, Hilden, Germany). Then exome sequence capture was performed on SureSelect Human All Exon V6 (Agilent). DNA library was subjected to 2×150 bp paired-end massively parallel sequencing using a Hiseq2000 Sequencing System (Illumina, San Diego, CA, USA). Before variant calling, sequence alignment files were generated to duplicate removal, local realignment around known Indels and base quality recalibration using the Genome Analysis Toolkit (GATK) (14). Variations that included single-nucleotide variants (SNVs) and insertions or deletions (Indels) were identified using both the VarScan 2.2.7 software package (http://www.ncbi.nlm.nih.gov/pubmed/22300766) (15) as well as the variant quality score recalibration (VQSR) protocol in GATK, and further filtered using a recommended threshold value (mapping quality >30, base quality >15, and read numbers >3). Then, SNP available at dbSNP130 (hg19) as well as those reported by the 1000 Genomes Project were filtered out from the output files using the ANNOVAR (http://nar.oxfordjournals.org/content/38/16/e164) (16).
After identifying a newly number of coding SNPs potentially associated with ARDS, SNP/InDel were tested by plink method to understand the difference between the outcome and subphenotypes of ARDS. While detecting the number and function of nonsynonymous SNV, the lung injury burden (LIB) was calculated by the ratio of nonsynonymous SNP number per megabase (MB) of DNA. The area under the receiver operating characteristic curve (ROC) was used for evaluating the predictive values of LIB in predicting outcome and subphenotype of patients with ARDS.
Statistical analysis
Data were presented as number (%) for categorical variables, and median (interquartile range) for continuous variables. Fisher’s exact test or χ2 test were used for categorical variables, and Student’s t-test or Mann-Whitney U test were used for continuous variables, as appropriate. The value of predictive ability was evaluated by the area under the curve (AUC) in the receiver operating characteristic (ROC) analysis. A P value <0.05 was considered statistically significant. Statistical analyses were carried out by the SPSS 16.0 software (IBM, Somers, NY, USA).
Results
There were 105 patients enrolled in the study, including 70 survivors and 35 non-survivors. The characteristics of outcome and subphenotype are presented in Table 1. The median age was 59 years old, while the median APACHE II score was 23, the median SOFA score and Murray lung injury score was 9 and 2.7, respectively. Among them, 91 patients were categorized as having pulmonary ARDS, 89 patients were diagnosed as sepsis and 66 patients as shock on enrollment.
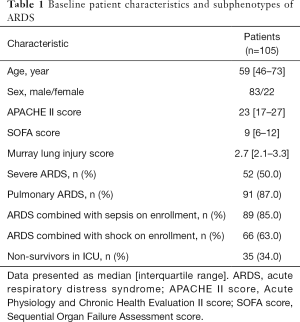
Full table
SNP/InDel data by whole-exome sequencing
By whole-exome sequencing, the number of SNP/InDel were 471,131 (Table 2). Among them, 120,830 SNP/InDel were in exonic region. The number of nonsynonymous SNV were 65,542, with 436 of frameshift-insertion for InDel and 897 of frameshift-deletion for InDel. GO analysis showed that 52 functions were correlated with ARDS development (P<0.01), and KEGG enrichment analysis showed that these SNP/InDel were in 10 pathways, such as cGMP-PKG signaling pathway, Platelet activation (P<0.05).
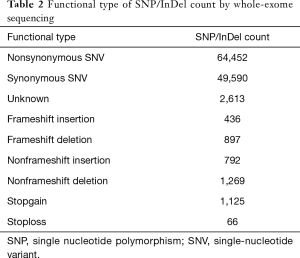
Full table
SNP/InDel data between ARDS patients with different outcome
LIB was tested to determine the quantitative differences between survivors and non-survivors. LIB of survivors was significantly higher than non-survivors [1,892 (1,848–1,942)/MB versus 1,864 (1,829–1,910)/MB, P=0.018]. GO and KEGG analysis were performed to determine the functional difference of SNP with outcome. GO analysis showed that 60 functions were correlated with ARDS outcome (P<0.01) (Figure 1), and KEGG enrichment analysis showed these SNP/InDel were in 13 pathways (Table 3), such as ECM-receptor interaction pathway, Platelet activation pathway and cGMP-PKG signaling pathway (P<0.01).

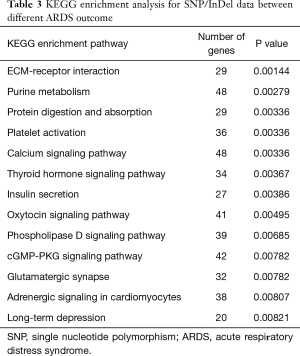
Full table
Association of genetic polymorphisms with ARDS outcome
To identify the novel SNPs which associated with ARDS outcome, the genotype distribution in different gene were summarized in Table 4, conformed to Hardy-Weinberg equilibrium. Although no strong evidence of stratification has been reported, several SNPs which potentially associated with ARDS outcome were found (Figure 2).
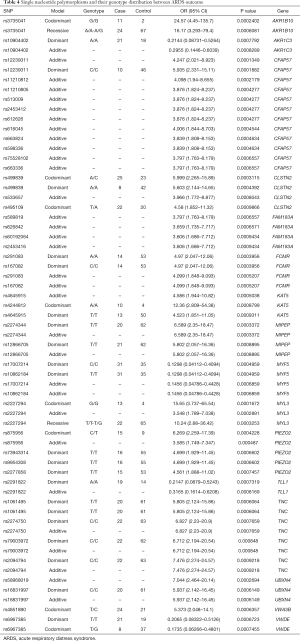
Full table

The value of LIB in predicting ARDS outcome
To determine the ability of LIB to predict ARDS outcome, analysis was carried out on LIB, P/F ratio, APACHE II score, SOFA score and Murray lung injury score with the area under the ROC curve of 0.6103 (P=0.0807), 0.568 (P=0.3124), 0.6763 (P=0.0053), 0.6204 (P=0.1002), 0.6614 (P=0.0581), respectively. The predicting value of LIB could increase to 0.712 (P=0.001) when combined with APACHE II score.
SNP/InDel data between ARDS patients with different subphenotypes
ARDS patients were divided into different subphenotypes (see online supplemental material in Appendix 1).
Severe ARDS group and non-severe group were divided according to the severity of lung injury. Compared with non-severe group, LIB was lower in severe ARDS group, with the ROC of predictive value of 0.727 (P<0.0001). GO analysis showed that 25 functions were correlated with ARDS severity (P<0.01), and KEGG enrichment analysis showed that these SNP/InDel were in 4 pathways, such as PI3K-Akt signaling pathway, ECM-receptor interaction (P<0.05).
ARDS patients were divided into pulmonary ARDS and extrapulmonary ARDS group. LIB was not significantly altered between the pulmonary and extrapulmonary ARDS. GO analysis showed that 19 functions were correlated with pulmonary and extrapulmonary ARDS (P<0.01), and KEGG enrichment analysis showed that these SNP/InDel were in 8 pathways, such as ECM-receptor interaction (P<0.05).
ARDS patients were divided into ARDS combined with sepsis and ARDS without sepsis on enrollment. Compared with patients without sepsis, the LIB was lower in ARDS combined with sepsis, with the ROC of predictive value of 0.6803 (P=0.0084). GO analysis showed that 24 functions were correlated with ARDS combined with sepsis (P<0.01), and KEGG enrichment analysis showed that these SNP/InDel were in 3 pathways, such as ECM-receptor interaction, Focal adhesion (P<0.05).
ARDS patients were divided into ARDS combined with shock and ARDS without shock on enrollment. Compared with patients without shock, the LIB was lower in ARDS combined with shock, with the ROC of predictive value of 0.6915 (P=0.0008). GO analysis showed that 46 functions were correlated with ARDS combined with shock (P<0.01), and KEGG enrichment analysis showed that these SNP/InDel were in 10 pathways, such as cAMP signaling pathway, ECM-receptor interaction (P<0.05).
Discussion
In this single-center, prospective observational trial which enrolled adult ARDS patients, whole exome-sequencing was performed to understand the difference between ARDS outcome and subphenotypes. The highlight of the study is the integrated framework of genetic variability of ARDS displayed through ARDS survivors and non-survivors. As defined by LIB, the mutational landscape of ARDS showed the overall genetic variability between survivors and non-survivors, while the detailed specific genetic polymorphisms which have an influence on outcome which finally showed genetic factors play a role in ARDS outcome.
As the role of genetics in the pathogenesis of ARDS is increasingly recognized, numerous genes and genetic variants were identified to proclaim their association with ARDS outcome. However, most were single genetic polymorphisms, little studies focus on the whole mutational landscape and its influence on ARDS outcome. To build an integrated framework, we classified different categories of genes, and try to observe their association with the outcome of ARDS, which are genes influencing immune regulation, genes influencing endothelial barrier function, genes influencing respiratory epithelial function, genes influencing coagulation, genes influencing injury and oxidative stress and so forth. However, as multiple different pathogenic processes, all these genes could interrelate.
Tumor mutation burden (TMB) is a marker which calculated as the nonsynonymous mutation number of per MB of DNA in tumor tissue (17). High TMB often correlates with a higher probability of tumor neoantigens, which could be recognized by lymphocytes (18,19), so it is hypothesized that the tumors with the highest TMB might be more likely to respond to immune checkpoint blockade therapy. Previous studies showed that patients with high TMB response better to immune checkpoint blockade therapy (20-22) and might have a better outcome (23). However, little data observed the mutation burden in ARDS, which might make a rough estimate on the whole mutational landscape of ARDS. In this study, we found when combined with clinical characteristics, burden could predict prognosis of ARDS.
We acknowledge some limitations in our study. Firstly, there was no validation group to study the association of functional SNPs with ARDS outcome which found by whole exome sequencing. Secondly, functional studies are needed to evaluate the mechanisms that underlie the associations between all the genetic variants and ARDS outcome and the mediating pathway. Thirdly, there was no healthy control group, the data were compared with reference genome UCSC hg19. In addition, the findings in the study were mainly pertinent to patients in single center who developed ARDS, and should be validated before generalization in cohorts.
Conclusions
Genetic variants are associated with ARDS outcome; however, their prognostic value still need to be verified by larger trials.
Acknowledgments
Funding: This work is partially supported by grants from the National Natural Science Foundations of China (81501705, 81571874, 81930058, 81671892, 81971888), grants from the Scientific Research Foundation of Graduate School of Southeast University (YBPY1604), grants from the Jiangsu Provincial Medical Youth Talent (QNRC2016808). This work is partially supported by grants from the Jiangsu Provincial Special Program of Medical Science (BL2013030, BE2018743, BE2019749), grants from Ministry of Science and Technology of China (2020YFC0843700), grants from Jiangsu Provincial Medical Priority Majors (ZDXKA2016025).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-5728
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-5728
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-5728). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study protocol was approved by the Ethics Committee (Approval Number: 2015ZDSYLL014.0) of Zhongda Hospital, School of Medicine, Southeast University, and written informed consent was obtained from each patient or their next of kin. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Fan E, Brodie D, Slutsky AS. Acute respiratory distress syndrome: advances in diagnosis and treatment. JAMA 2018;319:698-710. [Crossref] [PubMed]
- Bellani G, Laffey JG, Pham T, et al. Epidemiology, patterns of care, and mortality for patients with acute respiratory distress syndrome in intensive care units in 50 countries. JAMA 2016;315:788-800. [Crossref] [PubMed]
- Reilly JP, Christie JD. Linking genetics to ARDS pathogenesis: the role of the platelet. Chest 2015;147:585-6. [Crossref] [PubMed]
- Reilly JP, Christie JD, Meyer NJ. Fifty years of research in ARDS. Genomic contributions and opportunities. Am J Respir Crit Care Med 2017;196:1113-21. [Crossref] [PubMed]
- Grigoryev DN, Cheranova DI, Chaudhary S, et al. Identification of new biomarkers for Acute Respiratory Distress Syndrome by expression-based genome-wide association study. BMC Pulm Med 2015;15:95. [Crossref] [PubMed]
- Shortt K, Chaudhary S, Grigoryev D, et al. Identification of novel single nucleotide polymorphisms associated with acute respiratory distress syndrome by exome-seq. PLoS One 2014;9:e111953 [Crossref] [PubMed]
- Morrell ED, O'Mahony DS, Glavan BJ, et al. Genetic variation in MAP3K1 associates with ventilator-free days in Acute Respiratory Distress Syndrome. Am J Respir Cell Mol Biol 2018;58:117-25. [Crossref] [PubMed]
- Wei Y, Tejera P, Wang Z, et al. A Missense genetic variant in LRRC16A/CARMIL1 improves Acute Respiratory Distress Syndrome survival by attenuating platelet count decline. Am J Respir Crit Care Med 2017;195:1353-61. [Crossref] [PubMed]
- Dötsch A, Eisele L, Rabeling M, et al. Hypoxia Inducible Factor-2 Alpha and Prolinhydroxylase 2 Polymorphisms in patients with Acute Respiratory Distress Syndrome (ARDS). Int J Mol Sci 2017;18:1266. [Crossref] [PubMed]
- Sapru A, Liu KD, Wiemels J, et al. Association of common genetic variation in the protein C pathway genes with clinical outcomes in acute respiratory distress syndrome. Crit Care 2016;20:151. [Crossref] [PubMed]
- Kangelaris KN, Sapru A, Calfee CS, et al. The association between a Darc gene polymorphism and clinical outcomes in African American patients with acute lung injury. Chest 2012;141:1160-9. [Crossref] [PubMed]
- Moss M, Mannino DM. Race and gender differences in acute respiratory distress syndrome deaths in the United States: an analysis of multiple-cause mortality data (1979-1996). Crit Care Med 2002;30:1679-85. [Crossref] [PubMed]
- Erickson SE, Shlipak MG, Martin GS, et al. Racial and ethnic disparities in mortality from acute lung injury. Crit Care Med 2009;37:1-6. [Crossref] [PubMed]
- McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297-303. [Crossref] [PubMed]
- Koboldt D C, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012;22:568-76. [Crossref] [PubMed]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164 [Crossref] [PubMed]
- Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013;499:214-8. [Crossref] [PubMed]
- Matsushita H, Vesely MD, Koboldt DC, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature 2012;482:400-4. [Crossref] [PubMed]
- Maleki Vareki S. High and low mutational burden tumors versus immunologically hot and cold tumors and response to immune checkpoint inhibitors. J Immunother Cancer 2018;6:157. [Crossref] [PubMed]
- Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med 2017;377:2500-1. [Crossref] [PubMed]
- Cristescu R, Mogg R, Ayers M, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 2018;362:eaar3593 [Crossref] [PubMed]
- Chan TA, Yarchoan M, Jaffee E, et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol 2019;30:44-56. [Crossref] [PubMed]
- Devarakonda S, Rotolo F, Tsao MS, et al. Tumor mutation burden as a biomarker in resected non-small-cell lung cancer. J Clin Oncol 2018;36:2995-3006. [Crossref] [PubMed]


