
Related parameters of affinity and stability prediction of HLA-A*2402 restricted antigen peptides based on molecular docking
Introduction
Major histocompatibility complex class I (MHC-I) molecules plays an important role in the cellular immune response, presenting peptides to cytotoxic T lymphocytes (CTL) and allowing the immune system to carefully examine ongoing biological processes within the cell (1). Many studies on immunotherapy have found that tumor-specific antigen peptides not only bind to MHC by means of low affinity, but also often exhibit function defects in antigen processing and presentation, leading to immune evasion (2). This poses a huge challenge to T cell-based immunotherapy. HLA-A plays an important role in anti-tumor immune response and tumor neoantigen discovery, and HLA-A*24 is an allelic type of HLA-A (3,4). HLA-A*2402 is one of the most common alleles in East Asian populations, especially in Japanese and Chinese populations (5,6). Recent studies on HLA-A*2402 have focused on the clinical application of HLA-A*2402 restricted antigen. In a study on the safety, immune response rate and clinical benefit of cancer vaccine combined with chemotherapy, it was found that patients with HLA-A*2402 positive, locally advanced, metastatic and/or recurrent gastrointestinal, lung or cervical cancer, their specific T cell response rate of HLA-A*2402 restricted tumor-associated epitope peptide was significantly correlated with longer overall survival (7). In another study of new vaccine therapy evaluating HLA-A*2402 positive recurrent/progressive high-grade glioma patients using a variety of glioma cancer antigens and glioma angiogenesis-related antigen peptides, was found that this therapy was well tolerated, without any serious systemic adverse events, and could induce a strong antigen peptide-specific T lymphocyte response (8). However, the above-mentioned studies only used a variety of HLA-A*2402 restricted antigen peptides in combination with other therapeutic methods for anti-tumor therapy, and did not further explore how to replace HLA-A*2402 residues through molecular simulations, in order to better improve their roles in the anti-tumor immune response. Increasing the complementarity between the binding clefts of peptides and HLA-A molecules by replacing HLA anchor residues was a common step to improve the binding capacity and antigenicity of antigen peptides (9,10). However, this change must be based on the allelic types of each HLA-A molecule and may recruit different specific CTLs due to the conformational change of the antigen peptide, thereby reducing the recognition probability of T cells (11). The efficient presentation of polypeptide-MHC-I class (pMHC-I) molecular complexes to T cells benefited from the stable interaction between polypeptides and MHC-I (12). Compared with affinity, the stability of the pMHC-I complex could better reflect the immunogenicity of CTL (13), but it was difficult to distinguish stability from other elements of MHC-I binding, such as affinity. In recent years, scientists’ interest in artificial neural networks (ANN) has increased day by day. It is a rough simulation of the information processing capabilities of the human brain. It is a modern and complex computing technology that plays a huge role in drug analysis, drug technology, and screening of new drugs (14,15). Scientists have established a high-throughput method for evaluating the stability of the pMHC-I complexes using an ANN method to predict the half-life of pMHC-I complex binding (16). There are two novel tools that identify with relatively high accuracy. The two tools consist of (I) the NetMHCpan-4.1 server predicts binding of peptides to any MHC molecule of known sequence (17), and (II) NetMHCstab-1.0 predicts the stability of peptide binding to a number of different MHC molecules (18). Researches on molecular docking of protein-peptide interactions are difficult and time-consuming tasks because peptides are generally more flexible than proteins and tend to adopt multiple conformations. In the process of searching for binding sites for peptides, both peptide and protein molecules have significant conformational flexibility (19,20). At present, using the flexible molecular docking method in virtual screening to predict the binding affinity of polypeptides with different MHC allotypes has proven to have a fairly high prediction accuracy (21). The intent of the present study was to understand the intermolecular force, binding affinity, binding energy, affinity predicted values, and stability predicted values of HLA-A*2402 with restricted antigen peptides. In addition, we further analyzed the results of the in vitro competitive binding test, as well as the correlation between other parameters, and explored ways to improve the stability of the p-HLA-A*2402 complex.
Methods
Data collection
The polypeptide sequences of the HLA-A*2402 restricted antigen peptide and the results of the in vitro competitive binding test were obtained from the literature (22) and the affinity between the antigen peptide and HLA-A*2402 was predicted by the NetMHCpan v4.1 server (http://www.cbs.dtu.dk/services/NetMHCpan/) (17). The stability values of peptides and HLA-A*2402 were predicted by NetMHCstab v1.0 server (http://www.cbs.dtu.dk/services/NetMHCstab/) (18).
Molecular docking and dynamic simulation
The crystal model of the peptide-HLA-A*2402 complex (PDB ID: 2BCK) was obtained from the PDB database (http://www.rcsb.org/) (23,24) and Auto Vina (25) was used to dock the HLA-A*2402 restricted antigen peptide to the HLA-A*2402 protein in a flexible docking manner. Maestro (Schrodinger, LLC, New York, NY, 2019) (26) analyzed the intermolecular force of the complex [hydrogen bond (HB) and pi-stack] and the binding affinity values of the complex, while Flexpepdock (27) further optimized the docking morphology of the restricted antigen peptides and HLA-A*2402, and analyzed the binding affinity values of the complex. MM-GBSA (28) calculated the binding free energy values of HLA-A*2402-restricted antigen peptide. The above process was performed under the premise that the parameters of each docking system and the kinetic simulation were consistent.
Analysis of relevant parameters
The previous correlations of various parameters such as affinity prediction values, stability prediction values, intermolecular force, binding affinity, binding free energy, and in vitro competitive binding test results were analyzed and further explored the way to improve the affinity and stability of HLA-A*2402 with the restricted antigen peptides.
Statistical analysis
The correlation analysis used Spearman correlation coefficient statistical analysis, and P<0.05 was considered statistically significant.
Results
Relationship between the in vitro competitive binding capacity of peptides and the predicted values of affinity and stability
The sequences of HLA-A*2402 restricted antigen peptides, the results of the peptide in vitro competitive binding tests, the affinity predicted values of NetMHCpan v4.0, and the stability predicted values of NetMHCstab v1.0 are shown in Table 1.
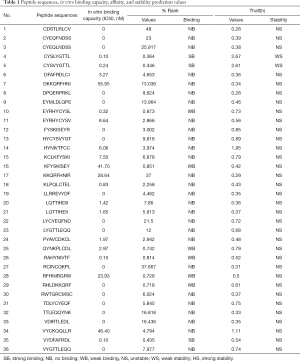
Full table
Among these, the threshold of affinity prediction: the threshold of strong binding prediction: %Rank ≤0.5 was identified as strong binding (SB), the threshold of weak binding prediction: 0.5< %Rank ≤2 (WB), and the rest of the values were identified as no binding (NB). Stability prediction threshold: strong stability (HS) prediction threshold (hours): Thalf(h) ≥6, weak stability (WS) prediction threshold (hours): 2≤ Thalf(h) <6, and other values were identified as unstable (NS). As shown in Table 1, 19 of the 36 antigen polypeptides were competitively binding with HLA-A*2402 molecules in vitro, and among these, three were predicted to have strong binding strength, five peptides were predicted to have weak binding capacity, and the remaining 11 were predicted to have no binding capacity. However, the remaining 17 polypeptides had no binding ability. Among the peptides with both in vitro competitive binding ability and predicted affinity, only two were predicted to have weak stability, namely CYSLYGTTL and CYSVYGTTL, respectively.
Intermolecular force and binding energy values of the peptide- HLA-A*2402 complex
The intermolecular forces (HB and pi-stack), binding affinity, and binding free energy values of the peptide-HLA-A*2402 complex are shown in Table 2. The intermolecular interaction that maintained the polypeptide-HLA-A*2402 complex are also shown in Table 2, and were mainly based on HB, followed by pi-stack.
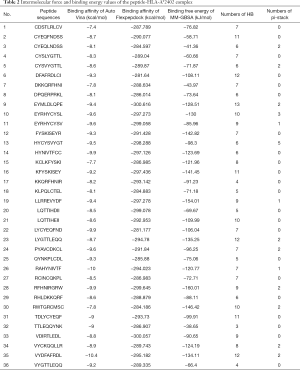
Full table
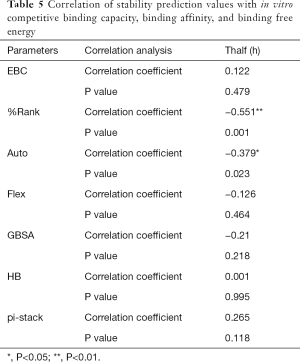
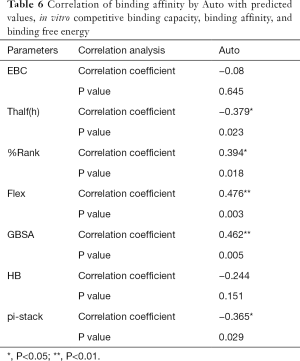
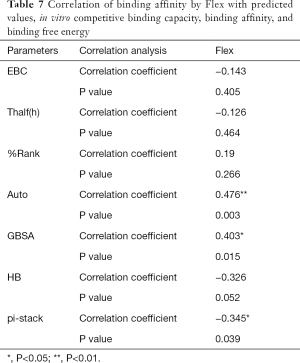
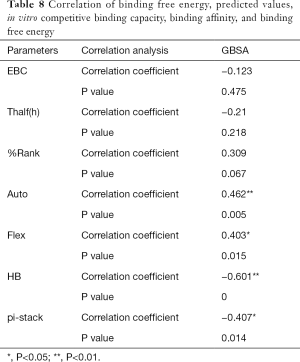
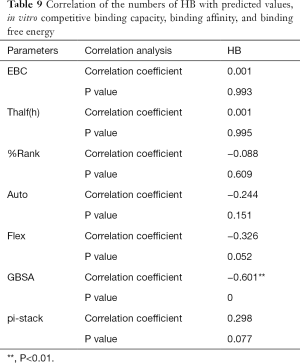
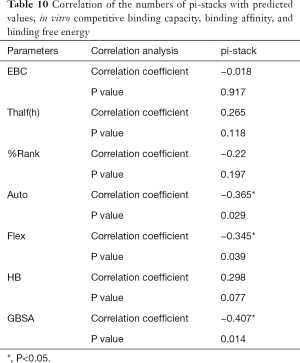
The correlation statistical analysis results of the in vitro competitive binding capacity (experimental binding capacity), the numbers of HB, the numbers of pi-stacks, the Auto binding capacity, the Flex binding capacity, the binding energy, the affinity prediction values (%Rank), and the stability prediction values [Thalf(h)] are shown in Tables 3-10, where * represented P<0.05, and ** represented P<0.01. Tables 3-10 show the results of correlation analysis among the Experimental binding capacity (EBC), the values of %Rank and Thalf(h), the Auto binding affinity, the Flex binding affinity, the GBSA binding free energy, and the numbers of HB and pi-stacks. The Spearman correlation coefficient was applied to indicate the strength of the correlation.
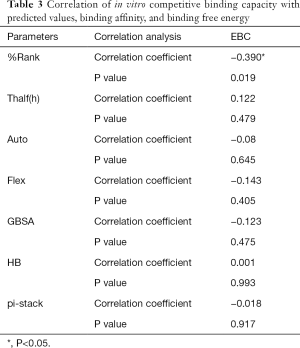
Full table
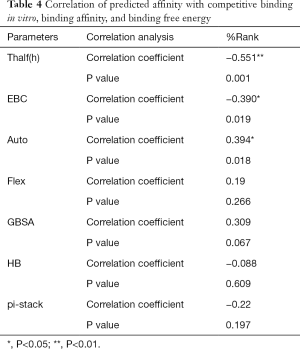
Full table
Full table
Full table
Full table
Full table
Full table
Full table
Table 3 shows that the correlation coefficient between EBC and %Rank was 0.623, and had a significant level of 0.01, indicating that there was a significant negative correlation between EBC and %Rank (P<0.05). However, there was no correlation among EBC and Thalf(h), Auto, Flex, and GBSA, numbers of HB and pi-stacks. Table 4 indicates that %Rank had a significant negative correlation with Thalf(h) and EBC (P<0.05), and a significant positive correlation with Auto (P<0.05), although there was no correlation among %Rank and Flex, GBSA, and the numbers of HB and pi-stacks. Table 5 shows that Thalf(h) had a significant negative correlation with %Rank and Auto (P<0.05), but there was no correlation among Thalf(h), EBC, Flex, GBSA, and the numbers of HB and pi-stacks. Table 6 shows that Auto had a significant positive correlation with %Rank, Flex, and GBSA (P<0.05), and a significant negative correlation with Thalf(h) and the numbers of pi-stacks (P<0.05). However, there was no correlation among Auto, EBC, and the numbers of HB. Table 7 shows Flex had a significant positive correlation with Auto and GBSA (P<0.05), and a significant negative correlation with the number of pi-stacks (P<0.05), but no correlation among Flex, EBC, Thalf(h), %Rank, and the numbers of HB seen. Table 8 shows that while GBSA had a significant positive correlation with Auto and Flex (P<0.05), and a significant negative correlation with the numbers of pi-stacks and HB (P<0.05), there was no correlation among GBSA, EBC, Thalf(h), and %Rank. Table 9 shows there is a significant negative correlation between the numbers of HB and GBSA (P<0.05). However, there was no correlation among the numbers of HB, EBC, Thalf(h), %Rank, Auto, Flex, and the numbers of pi-stacks. Finally, Table 10 shows there is a significant negative correlation among the number of pi-stacks, Auto, Flex, and GBSA (P<0.05), but no correlation among the numbers of pi- stacks, EBC, Thalf(h), %Rank, and the numbers of HB.
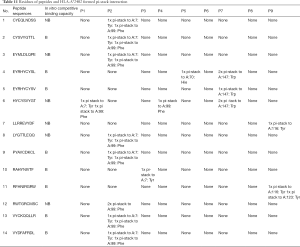
Binding affinity by Auto was related to the predicted values of affinity and stability, binding affinity by Flex, and binding free energy. Moreover, it also closely related to the intermolecular force pi-stack. There was a significant negative correlation among the numbers of pi-stacks, binding affinity, and binding free energy. This suggests that the numbers of pi-stacks played an important role in the interaction of peptides and HLA-A*2402. Furthermore, the amino acid residues that form the pi-stack interaction between the polypeptide and HLA-A*2402 were screened, as shown in Table 11, which also shows P1-P9 represented the amino acid residues 1 to 9 of the polypeptide, respectively, and A represented HLA-A*2402. The results also indicate that pi-stacks were mainly composed of Y (Tyr) on the polypeptide, which was mainly located at position 2, 4, 5, and 7, H (His) that was mainly located at position 1 and 3, W (Trp) that was located at position 2 and 9, and F (Phe) that was located at position 9. However, the residues of position 6 and 8 of polypeptides did not form a pi-stack with HLA-A*2402. The residues on HLA-A*2402 that formed pi-stacks with those on the polypeptide were mainly Y (Tyr) at positions 7, 116, and 123, H (Hie) at position 70, F (Phe) at position 99, and W (Trp) at position 147. It can also be seen that the interaction of pi-stacks mainly occurred among residues Y, H, W, and F, and H was a positively charged basic amino acid, while Y, W, and F were all aromatic amino acids.
Full table
Residues replacement, and affinity and stability values prediction
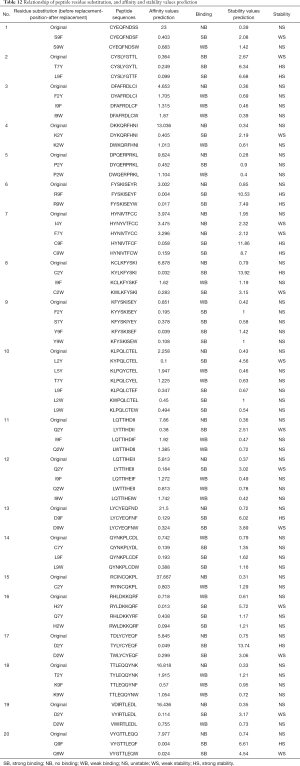
Certain residues on polypeptides that did not form pi-stacks with HLA-A*2402 were replaced as follows: replacing the non-Y (Tyr) at position 2, 4, 5, and 7 of polypeptide with Tyr; the non-H (His) at position 1 and 3 with His; the non-W (Trp) at position 2 and 9 with Trp; and the non-F (Phe) at position 9 with Phe. Table 12 shows the predicted values of affinity and stability of the polypeptide before and after residue replacement with HLA-A*2402. It is seen that after the residue replacement, of the 22 polypeptides that did not produce intermolecular pi-stack interaction with HLA-A*2402 before, 20 polypeptides had improved affinity and/or stability after the residue replacement. In addition, when excluding the residue substitution sites with affinity and/or stability reduced or unchanged, it was found that the residue sites that mainly occurred at position 2 (C2Y, F2Y, Q2Y, H2Y, L2Y, P2Y, T2Y, D2W, Q2W, H2W, L2W, P2W), position 4 (I4Y), position 7 (T7Y, C7Y, Q7Y), and position 9 (C9F, D9F, K9F, Q9F, S9F, Y9F, C9W, D9W, K9W, Q9W, S9W, Y9W). This means the residues (C, D, Q, H, L, P, T) at position 2 on the polypeptide were replaced by Y and W, the residues (I) at position 4 were replaced by Y, the residues (T, C, Q) at position 7 were replaced by Y, and the residues (C, D, K, Q, S, Y) at position 9 were replaced by F and W, and the predicted affinity and/or stability values of the polypeptide after the replacement were all higher than before. At the same time, it was found that the predicted strong/weak affinity between HLA-A*2402 and the peptide did not necessarily mean that they had strong/weak stability, while the complexes predicted to have strong/weak stability must have strong/weak affinity between the molecules.
Full table
Discussion
Polypeptides in the MHC peptide binding groove have been shown to mediate the recognition of T cells and other receptors by affecting the binding function of the complex. Peptides could regulate the movement of MHC itself, thereby prompting the recognition of the peptide-MHC complex by other receptors. Structural modeling of the peptide-MHC complex may reveal unknown driving factors for T cell activation, thereby contributing to the development of better and safer immunotherapy (29). In the present study, we found the intermolecular interactions of the polypeptide-HLA-A*2402 complex were maintained mainly by HB, followed by pi-stack. The binding affinity calculated by molecular docking also showed a significant negative correlation with the intermolecular force pi-stack and the pi-stack had a significant negative correlation with the binding affinity and binding free energy. The residues (C, D, Q, H, L, P, T) at position 2 on the polypeptide that did not form the intermolecular pi-stack force with HLA-A*2402 were replaced by Y and W, the residue (I) at position 4 was replaced by Y, the residues (T, C, Q) at position 7 were replaced by Y, the residues (C, D, K, Q, S, Y) at position 9 were replaced by F and W, and the predictive values of affinity and/or stability were improved when compared to the previous replacement.
Current studies have shown that the substitution of proline (Pro) for the third residue on the polypeptide could not only significantly enhance the ability of anti-tumor CTL to recognize wild-type epitopes (30), but also increase the affinity of pMHC and the stability of the complex (31). After analyzing the crystal structure of the MHC-peptide complex, the conformation of the modified antigen polypeptide was found to be like the wild type, and it interacted with the most conserved tyrosine residue Y159 in mammalian MHC-I molecules and maintained a stable bond (32). Changes in the identity of anchor residues may have significant effects, such as changing the conformation of the peptide-MHC complex, thereby affecting contact between the residues on the polypeptides and TCRs. Binding of the TCR-like recombinant antibody to the melanoma differentiation antigen gp100 T cell epitope G9-209 were completely dependent on the identity of the second single peptide anchor residue. In other words, the TCR-like antibody could only be modified with high affinity to HLA-A2 peptide G9-209-2M and then be recognized by specific complexes after contacting. It was suggested that the modification of anchor residues could significantly affect the conformation of the MHC peptide groove, which may have a profound impact on the interaction of TCR-pMHC molecules (33,34). Compared with non-antigenic peptides, antigenic peptides tend to bind to MHC-I molecules more stably, and results confirm that the unsuitable anchor residue at position 2 of the polypeptide is particularly prone to unstable interaction with MHC-I (13). The in vitro competitive binding ability after residue substitution at the above sites still requires further clarification in in vitro tests, and we will perform the Enzyme-Linked Immunosorbent spot (ELISpot) assay (35), immune repertoire (36,37) and peptide-MHC tetramer staining (38) to verify the prediction results, moreover, other factors that affect the affinity and stability of the polypeptides with HLA-A*2402 will require multidisciplinary, multidimensional analysis and discussion.
Conclusions
The generation and increase in the numbers of pi-stack interactions between antigen peptides and HLA-A*2402 may help improve the affinity and stability of the complex. The prediction of peptide-HLA intermolecular force and binding affinity by means of molecular docking is a supplement to the current commonly used prediction databases.
Acknowledgments
Funding: The study was supported by the Youth Project of National Natural Science Foundation of China (No. 82004129) and the Sci-Tech Key Project of Zhejiang Province of China (No. 2017C03053).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-630). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Pedersen LE, Harndahl M, Rasmussen M, et al. Porcine major histocompatibility complex (MHC) class I molecules and analysis of their peptide-binding specificities. Immunogenetics 2011;63:821-34. [Crossref] [PubMed]
- Zou W. Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nat Rev Cancer 2005;5:263-74. [Crossref] [PubMed]
- Carreno BM, Magrini V, Becker-Hapak M, et al. Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science 2015;348:803-8. [Crossref] [PubMed]
- Berzofsky JA, Terabe M, Trepel JB, et al. Cancer vaccine strategies: translation from mice to human clinical trials. Cancer Immunol Immunother 2018;67:1863-9. [Crossref] [PubMed]
- Sobao Y, Sugi K, Tomiyama H, et al. Identification of hepatitis B virus-specific CTL epitopes presented by HLA-A*2402, the most common HLA class I allele in East Asia. J Hepatol 2001;34:922-9. [Crossref] [PubMed]
- Nakaoka H, Inoue I. Distribution of HLA haplotypes across Japanese Archipelago: similarity, difference and admixture. J Hum Genet 2015;60:683-90. [Crossref] [PubMed]
- Murahashi M, Hijikata Y, Yamada K, et al. Phase I clinical trial of a five-peptide cancer vaccine combined with cyclophosphamide in advanced solid tumors. Clin Immunol 2016;166-167:48-58. [Crossref] [PubMed]
- Kikuchi R, Ueda R, Saito K, et al. A Pilot Study of Vaccine Therapy with Multiple Glioma Oncoantigen/Glioma Angiogenesis-Associated Antigen Peptides for Patients with Recurrent/Progressive High-Grade Glioma. J Clin Med 2019;8:263. [Crossref] [PubMed]
- Abella JR, Antunes D, Jackson K, et al. Markov state modeling reveals alternative unbinding pathways for peptide-MHC complexes. Proc Natl Acad Sci U S A 2020;117:30610-8. [Crossref] [PubMed]
- Abella JR, Antunes DA, Clementi C, et al. Large-Scale Structure-Based Prediction of Stable Peptide Binding to Class I HLAs Using Random Forests. Front Immunol 2020;11:1583. [Crossref] [PubMed]
- van der Burg SH, Arens R, Ossendorp F, et al. Vaccines for established cancer: overcoming the challenges posed by immune evasion. Nat Rev Cancer 2016;16:219-33. [Crossref] [PubMed]
- Lebedeva T, Anikeeva N, Kalams SA, et al. Major histocompatibility complex class I-intercellular adhesion molecule-1 association on the surface of target cells: implications for antigen presentation to cytotoxic T lymphocytes. Immunology 2004;113:460-71. [Crossref] [PubMed]
- Harndahl M, Rasmussen M, Roder G, et al. Peptide-MHC class I stability is a better predictor than peptide affinity of CTL immunogenicity. Eur J Immunol 2012;42:1405-16. [Crossref] [PubMed]
- Barrett DG, Morcos AS, Macke JH. Analyzing biological and artificial neural networks: challenges with opportunities for synergy? Curr Opin Neurobiol 2019;55:55-64. [Crossref] [PubMed]
- Wesolowski M, Suchacz B. Artificial neural networks: theoretical background and pharmaceutical applications: a review. J AOAC Int 2012;95:652-68. [Crossref] [PubMed]
- Rasmussen M, Fenoy E, Harndahl M, et al. Pan-Specific Prediction of Peptide-MHC Class I Complex Stability, a Correlate of T Cell Immunogenicity. J Immunol 2016;197:1517-24. [Crossref] [PubMed]
- Reynisson B, Alvarez B, Paul S, et al. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res 2020;48:W449-W454. [Crossref] [PubMed]
- Jørgensen KW, Rasmussen M, Buus S, et al. NetMHCstab - predicting stability of peptide-MHC-I complexes; impacts for cytotoxic T lymphocyte epitope discovery. Immunology 2014;141:18-26. [Crossref] [PubMed]
- Agrawal P, Singh H, Srivastava HK, et al. Benchmarking of different molecular docking methods for protein-peptide docking. BMC Bioinformatics 2019;19:426. [Crossref] [PubMed]
- Kurcinski M, Badaczewska-Dawid A, Kolinski M, et al. Flexible docking of peptides to proteins using CABS-dock. Protein Sci 2020;29:211-22. [Crossref] [PubMed]
- Bordner AJ, Abagyan R. Ab initio prediction of peptide-MHC binding geometry for diverse class I MHC allotypes. Proteins 2006;63:512-26. [Crossref] [PubMed]
- Bonsack M, Hoppe S, Winter J, et al. Performance Evaluation of MHC Class-I Binding Prediction Tools Based on an Experimentally Validated MHC-Peptide Binding Data Set. Cancer Immunol Res 2019;7:719-36. [Crossref] [PubMed]
- Cole DK, Rizkallah PJ, Gao F, et al. Crystal structure of HLA-A*2402 complexed with a telomerase peptide. Eur.J.Immunol 2006;36:170-9. [Crossref] [PubMed]
- Burley SK, Berman HM, Bhikadiya C, et al. RCSB Protein Data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res 2019;47:D464-D474. [Crossref] [PubMed]
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010;31:455-61. [PubMed]
- Özçelik AB, Özdemir Z, Sari S, et al. A new series of pyndazinone derivatives as chohnesterases inhibitors: Synthesis, in vitro activity and molecular modeling studies. Pharmacol Rep 2019;71:1253-63. [Crossref] [PubMed]
- London N, Raveh B, Cohen E, et al. Rosetta FlexPepDock web server - high resolution modeling of peptide-protein interactions. Nucleic Acids Res 2011;39:W249-53 [Crossref] [PubMed]
- Wang E, Sun H, Wang J, et al. End-Point Binding Free Energy Calculation with MM/PBSA and MM/GBSA: Strategies and Applications in Drug Design. Chem Rev 2019;119:9478-508. [Crossref] [PubMed]
- Ayres CM, Corcelli SA, Baker BM. Peptide and Peptide-Dependent Motions in MHC Proteins: Immunological Implications and Biophysical Underpinnings. Front Immunol 2017;8:935. [Crossref] [PubMed]
- Antunes DA, Abella JR, Devaurs D, et al. Structure-based Methods for Binding Mode and Binding Affinity Prediction for Peptide-MHC Complexes. Curr Top Med Chem 2018;18:2239-55. [Crossref] [PubMed]
- Hafstrand I, Doorduijn EM, Sun R, et al. The Immunogenicity of a Proline-Substituted Altered Peptide Ligand toward the Cancer-Associated TEIPP Neoepitope Trh4 Is Unrelated to Complex Stability. J Immunol 2018;200:2860-8. [Crossref] [PubMed]
- Uchtenhagen H, Abualrous ET, Stahl E, et al. Proline substitution independently enhances H-2Db complex stabilization and TCR recognition of melanoma-associated peptides. Eur J Immunol 2013;43:3051-60. [Crossref] [PubMed]
- van Stipdonk MJ, Badia-Martinez D, Sluijter M, et al. Design of Agonistic Altered Peptides for the Robust Induction of CTL Directed towards H-2Db in Complex with the Melanoma-Associated Epitope gp100. Cancer Res 2009;69:7784-92. [Crossref] [PubMed]
- Denkberg G, Klechevsky E, Reiter Y. Modification of a Tumor-Derived Peptide at an HLA-A2 Anchor Residue Can Alter the Conformation of the MHC-Peptide Complex: Probing with TCR-Like Recombinant Antibodies. J Immunol 2002;169:4399-407. [Crossref] [PubMed]
- Slota M, Lim JB, Dang Y, et al. ELISpot for measuring human immune responses to vaccines. Expert Rev Vaccines 2011;10:299-306. [Crossref] [PubMed]
- Liu X, Wu J. History, applications, and challenges of immune repertoire research. Cell Biol Toxicol 2018;34:441-57. [Crossref] [PubMed]
- Schober K, Buchholz VR, Busch DH. TCR repertoire evolution during maintenance of CMV-specific T-cell populations. Immunol Rev 2018;283:113-28. [Crossref] [PubMed]
- Newell EW, Sigal N, Nair N, et al. Combinatorial tetramer staining and mass cytometry analysis facilitate T-cell epitope mapping and characterization. Nat Biotechnol 2013;31:623-9. [Crossref] [PubMed]
(English Language Editor: B. Draper)


