
Integrated genomic and transcriptomic analysis suggests KRT18 mutation and MTAP are key genetic alterations related to the prognosis between astrocytoma and glioblastoma
Introduction
Glioma is the most common tumor of the brain. Approximately 25.5% of all primary brain and other central nervous system (CNS) tumors and 80.8% of malignant brain tumors are glioma (1). The 2016 World Health Organization Classification of Tumors of the Central Nervous System (CNS WHO, 2016) classifies glioma into grade I to grade IV (2). Glioblastoma (GBM) is classified as grade IV and is the most malignant among all CNS tumors (1). The prognosis of GBM patients is poor, with a 5-year survival rate of less than 5% (3,4). Tumor heterogeneity and the ambiguity of histomorphology are the two main challenges to the diagnosis and treatment of GBM.
Astrocytoma, another subtype of glioma, is characterized by isocitrate dehydrogenase (IDH1) or (IDH2) mutation, which can be accompanied by TP53 and ATRX mutation (5). According to the status of the IDH gene, astrocytoma can be divided into 3 subtypes: IDH mutant, IDH wild type, and not otherwise specified (NOS). Among them, the IDH mutation subtype has the potential to progress to GBM (6). Histologically, astrocytoma can be divided into diffuse astrocytoma (DA) and anaplastic astrocytoma (AA). DA, characterized by high differentiation and slow growth, is classified as low-grade glioma (grade II), while AA is classified as high-grade glioma (grade III) due its high proliferative activity (7,8). Both molecular and histological characteristics are important to the prognosis of astrocytoma. AA patients with the IDH mutation often have good prognosis, while AA patients with wild-type IDH1 have a poor prognosis and high risk of progression. There is no significant prognostic difference between AA with IDH wild type and GBM with IDH wild type (9). Therefore, the same treatment protocol for GBM patients is recommended for patients with AA wild type IDH if they also have a positive MGMT methylation status (10). Due to astrocytoma's poor prognosis and high risk of progression to GBM, it is important to explore the molecular characteristics of astrocytoma.
In this study, we collected tumor samples from 12 patients with astrocytoma and 12 patients with GBM, and performed whole-exome sequencing (WES) and RNA sequencing (RNA-seq) analysis on them to compare the genomic and transcriptomic characterization between these 2 tumors. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-21-1317).
Methods
Patients and samples
In total, 24 patients with glioma, including 12 with astrocytoma and 12 with GBM, were recruited at Nanfang Hospital, Southern Medical University from MAR 2019 to AUG 2020. All procedures involving human participants were in accordance with principles of the Declaration of Helsinki (2013). All patients signed informed consent, and the study design was approved by the Ethics Committee of Nanfang Hospital (NFEC-2019-006). Tumor tissue and matched normal blood were collected from each patient. Hematoxylin and eosin (HE) stained histological sections of tumors were reviewed by an expert pathologist to ensure that the tumor content was above 20%. Clinical records were obtained from the electronic medical records of the hospital.
WES
Maxwell RSC blood DNA kit (cat no. AS1400, Promega) was used to extract DNA in snap-frozen tissue and peripheral whole blood, and MagMAX FFPE DNA/RNA Ultra kit (cat no A31881, Thermo Fisher Scientific) was used to extract DNA in formalin-fixed, paraffin-embedded (FFPE) tissue. A Covaris LE220 sonicator was used for shearing with purified DNA. Probes from the SureSelect XT Human All Exon V7 kit (cat. no. 5991–9039, Agilent) were used to hybridize with DNA fragments. After being polymerase chain reaction (PCR)—amplified and end-repaired, captured DNA was attached to the adapters and barcode using a SureSelect XT HS and Low Input Library Preparation Kit for ILM (pre PCR) kit (cat. no. G9704, Agilent). Libraries were sequenced on an Illumina NovaSeq-6000 sequencer (Illumina). The reads were aligned to hg19 after removing adapters and low-quality reads using the Burrows-Wheeler Alignment algorithm and the Genome Analysis Toolkit (GATK, version 3.5).
RNA-seq
The MagMAX FFPE DNA/RNA Ultra kit (Thermo Fisher Scientific) was used to extract RNA from FFPE tissue. The NEBNext RNA First Strand Synthesis Module [cat. no. E7525S, New England Biolabs Inc. (NEB)] was used for reverse transcription, and the NEBNext Ultra II Non-Directional RNA Second Strand Synthesis Module (cat. no. E6111S, NEB) was used for complement DNA (cDNA) synthesis. SureSelect XT HS and Low Input Library Preparation Kit for ILM (Pre PCR) (cat. no. G9704, Agilent) was used for RNA-seq library preparation. The libraries were sequenced on an NovaSeq-6000 Sequencing System (Illumina, USA). Although it is a difficult to use FFPE samples for RNA-seq, studies have established reliable protocols (11,12). NEBNext RNA First Strand Synthesis Module and NEBNext Ultra II Non-Directional RNA second Strand Synthesis Module, 2 commercial kits which were developed from the protocols, were used for RNA-seq in this study.
Differential gene expression analysis
STAR (version 020201) was used to align the reads to the reference genome (hg19), and StringTie2 (version 1.3.5) was used for assembly. After normalization was completed using upper quartile counts, a list of differentially expressed genes between astrocytoma and GBM was compiled using DESeq2. An adjusted P value <0.1 and an absolute value of Log2 fold change >1 was set as the significance threshold.
Immune cell infiltration analysis
The infiltration of immune cells was analyzed by the CIBERSORT Algorithm with LM22 immune subsets (13). The relative scores of 22 human immune cell types were quantified using 547 marker genes with a gene-based deconvolution algorithm. The standardized data set was uploaded to the CIBERSOFT website (https://cibersort.stanford.edu/index.php) and run using 1,000 aligned default signature matrices.
Statistical analysis
GraphPad Prism 8 (GraphPad Software, Inc., CA, USA) and R version 4.0.3 (R Foundation for Statistical Computing) were used for statistical analyses and data visualization. Unless stated otherwise, an unpaired t-test was used in all group comparisons, and a P value <0.05 was considered significant.
Results
Clinicopathological and epidemiological information of patients
Astrocytoma and GBM are the 2 main subtypes of glioma. Exploring the molecular characteristics of these 2 subtypes have a great value for clinical diagnosis and treatment. In this study, 12 patients with GBM and 12 patients with astrocytoma were enrolled and WES and RNA-seq were performed. The overall clinical data of these patients are shown in Table 1. There were 14 male patients and 10 female patients (P=0.214). The median age was 45.5 (range, 18–68) years old, with the age of GBM patients being greater than that of astrocytoma patients (P=0.048). There were 7 patients with IDH1 mutation and 17 patients with IDH1 wild type. Among the patients with IDH1 mutation, there were 6 patients with astrocytoma and only 1 patient with GBM (P=0.068), indicating that the majority of GBM patients in this study had primary GBM. In addition, the methylation of the MGMT promoter and the mutation of the TERT promoter were detected in 24 patients. There were 10 patients with MGMT-promoter methylation, 5 of whom had GBM and 5 of whom had astrocytoma (P=0.640). There were 11 patients with a mutation in the TERT promoter, including 10 patients with GBM and 1 patient with astrocytoma (P<0.001).
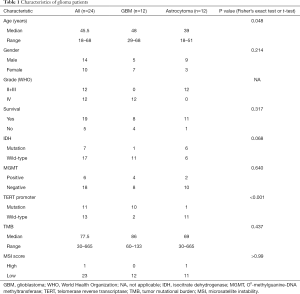
Full table
Somatic mutation profile in astrocytoma and GBM
WES was performed on the 24 patients, and the somatic mutation gene was analyzed. The top 20 mutated genes are shown in Figure 1A. TP53, FRG1, and IDH1 were the top 3 mutated genes in glioma, and their mutation rates were 54% (13/24), 33% (8/24), and 29% (7/24), respectively. The mutation rates of several genes differed between astrocytoma and GBM. IDH1 mutations were found in 50% (6/12) of astrocytoma cases but in only 8% (1/12) of GBM cases. Usually, the IDH1 mutation of GBM progresses from astrocytoma with the IDH1 mutation, and is called secondary GBM (14). In addition, TP53, KRT18, and ATRX were enriched in astrocytoma, but TTN was enriched in GBM (Figure 1A). Statistical analysis showed that KRT18 was the only gene which was significantly enriched in astrocytoma (Fisher’s exact test, P=0.037; Figure 1B).
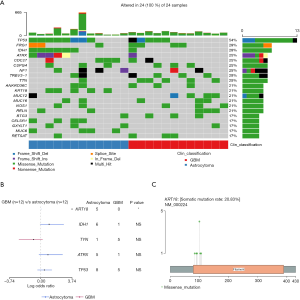
Next, we analyzed the mutation sites of the KRT18 gene in the patients; 6 different mutation sites were found in 5 patients with astrocytoma, but none were found in GBM patients. The most frequent mutation was p.S100R, found in 4 out of the 5 patients. The other mutations included p.T103N, p.R106W, p.M84I, p.R90C, and p.A92P (Figure 1C). All the mutation sites were located in the Coil 1A domain [80–115] of the KRT18 gene. Studies have reported that this region is closely related to the interaction of tumor necrosis factor (TNF) receptor 1-associated protein (TRADD) and the pinin protein (15,16). TRADD is a tumor suppressor associated with cell apoptosis, while pinin is involved in the regulation of cell adhesion and modulation of the activity of multiple tumor suppressor genes (17,18). The above results showed that somatic mutation of KRT18 gene in astrocytoma may affect the apoptosis and adhesion of glioma cells.
Copy number variations (CNVs) in astrocytoma and GBM
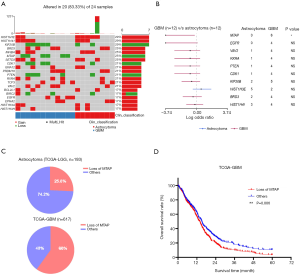
A CNV is a type of duplication or deletion event that affects a large stretch of base pairs. We analyzed the copy number amplification or deletion of genes in the patients. The results revealed amplification at HIST1H3E, HIST1H4I, BRD3, INHBA, RXRA, and SETD3 loci, and deletions at KIF20B, MTAP, CDK1, PTEN, and BIRC2 loci (Figure 2A). Copy number loss of MTAP was the only event significantly enriched in GBM (Fisher’s exact test, P=0.014; Figure 2B). To verify this result, we analyzed the frequency of the copy number loss of MTAP in astrocytoma and GBM by using The Cancer Genome Atlas Low Grade Glioma (TCGA-LGG) data and TCGA-GBM data. Results showed that 25.8% of TCGA patients with astrocytoma have copy number loss of the MTAP gene, but the proportion reached 60% in GBM (Figure 2C). This was consistent with the results of our study. We further analyzed the prognosis of patients from the TCGA-GBM data set. Results showed that the survival time of patients with copy number loss of the MTAP gene was shorter than that in other patients (P=0.006; Figure 2D). These results suggested that MTAP may be a tumor suppressor gene and that loss of MTAP is closely related to a poor prognosis in glioma.
Differential patterns of somatic copy number alterations (SCNAs) in astrocytoma and GBM
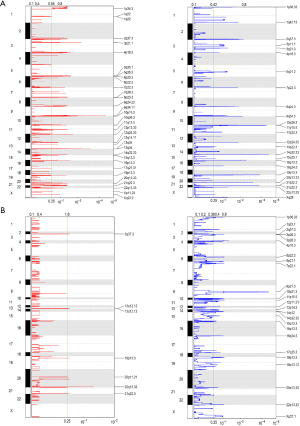
To further analyze the SCNAs in the astrocytoma and GBM patients, GISTIC2.0 was used to detect SCNA. In the astrocytoma patients, 2 amplification peaks were found at cytobands 9q34.11 and 13q14, and several regions of chromosomal loss were found at cytobands 14q32.33, 11p15.5, 19p13.3, and 21q22.3 (Figure 3A). Meanwhile, no amplification peak was found in the GBM patients, and only 1 region of chromosomal loss at 9p21.3 was discovered, which includes several tumor suppressor genes, such as MTAP, CDKN2A, and, CDKN2B (Figure 3B). An earlier study reported that low CDKN2B expression and loss of 9p21.3 were associated with worse prognosis in GBM (19). Taken together, these results indicated that the SCNA pattern differs between astrocytoma and GBM.
Differential signal pathway enrichment in astrocytoma and GBM
To explore the differences in the transcriptome between astrocytoma and GBM, RNA-seq was performed in the 24 patients. Principal component analysis (PCA) showed that astrocytoma and GBM had distinct gene expression profiles (PC1 =22%, PC2 =18%; Figure 4A). Differentially expressed genes were analyzed using DESeq2, and the significance threshold was set at adjusted P value <0.1 with an absolute value of Log2 fold change (LogFC) >1 (Figure 4B). Gene Ontolgoy (GO) biological process analysis was performed in upregulated genes and downregulated genes, and the top 10 pathways are shown in Figure 4C,D. The results showed that the upregulated genes in GBM were mainly enriched in tumor-related pathways, such as cell adhesion, cell division, and angiogenesis (Figure 4C). However, the upregulated genes in astrocytoma were mainly enriched in pathways of ion transport, cell-cell signaling, and neurogenesis (Figure 4D). The above results indicated that GBM may have higher cell proliferation and invasion ability when compared with astrocytoma.
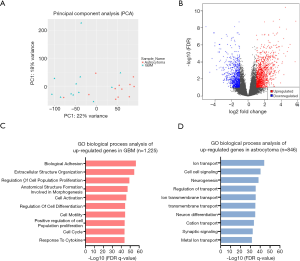
RNA-seq revealed differentially expressed genes between astrocytoma and GBM
A total of 1,242 genes were upregulated and 812 genes were downregulated in GBM when compared with astrocytoma. A heatmap was constructed to show the top 50 differentially expressed genes (Figure 5A and Table S1). RELN was the most significantly enriched gene in astrocytoma when compared with GBM [logFC =5.08 (astrocytoma over GBM); adjusted P value =7.38e-09; Figure 5A and Table S1]. RELN encodes a large secreted extracellular matrix protein which regulates the cell positioning and neuronal migration during brain development (20). It can modulate the growth and substrate-dependent migration of glioma cells (21), with downregulation of RELN predicting a poor prognosis in glioma (22).
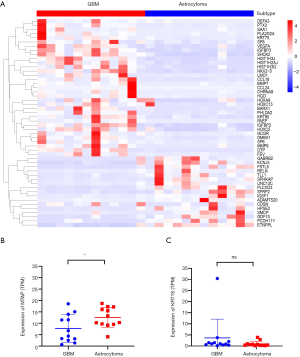
In addition, we analyzed the differentially expressed genes which were upregulated in GBM, with OTP being found to be the highest rank gene [logFC =5.78 (GBM over astrocytoma); adjusted P value=1.66e-09; Figure 5A and Table S1]. It is involved in brain development and is a prognostic hypoxia-associated gene in IDH-mutant glioma (23). CCL18, a small cytokine of the CC chemokine family, was highly expressed in GBM when compared with astrocytoma (logFC =5.08, adjusted P value =8.88e-07; Figure 5A and Table S1). It is produced and secreted mainly by the innate immune system (24). Studies have found that CCL18 is highly expressed in glioma and other CNS patients with traumatic brain injuries or neoplastic disorders (25,26), and that CCL18 is an immune-related risk signature for GBM and can promote the invasion of GBM cells (27).
We further found several genes associated with cell migration and proliferation, including MMP7, MMP9, IGFBP2, and IGGBP3. These results were consistent with the characteristics of GBM that are presented in Figure 4C. Cumulatively, these findings indicated that astrocytoma and GBM have distinct gene expression patterns.
Our results above (Figure 2) showed that copy number loss of the MTAP gene was significantly enriched in GBM, but was absent in astrocytoma. It would be of interest to see whether its expression is consistent with the copy number alterations obtained from the WES data. Indeed, the expression of MTAP in GBM obtained from RNA-seq was significantly lower than that in astrocytoma (Figure 5B). We also analyzed the expression of KRT18 in astrocytoma and GBM but found no difference between the two (Figure 5C).
Infiltration of immune cells in astrocytoma and GBM
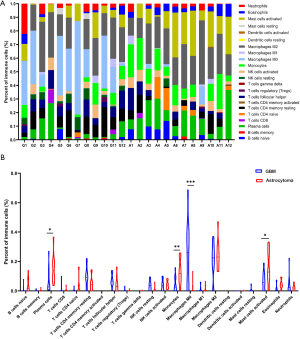
Immune checkpoint inhibitors (ICIs) have become an area of intense research in glioma, but the objective response rate of ICIs in glioma is only about 10%. Tumor microenvironment (TME) may be one of the more important factors which affect the response to ICIs. Therefore, CIBERSORT was used to analyze the RNA-seq data and assess the infiltration of 22 immune cell types in the tumor microenvironment of the patients (13). The results showed that most immune cell types, including cluster of differentiation (CD)8+ T cells, B cells, CD4+ T cells, and M1 macrophages, were missing from the microenvironment of the patients (Figure 6A and Table S2). Although M2 macrophages were enriched in the TME, there was no significant difference between astrocytoma and GBM (Figure 6B). However, we found that M0 macrophages were significantly enriched in GBM, while activated mast cells, monocytes, and plasma cells were significantly enriched in astrocytoma (Figure 6B). These immune cells may closely relate to the formation of the microenvironment in astrocytoma and GBM.
Discussion
According to the histologic criteria of CNS WHO (2016), DA is categorized as grade II, and AA is categorized as grade III (2). However, multiple studies have shown that diffuse or AA with IDH wild type has an aggressive clinical course and poor prognosis, similar to patients with IDH wildtype GBM (9,28,29). The classification and grading of IDH wildtype astrocytoma may not accurately reflect the molecular pathogenesis and clinical outcomes of this tumor type. Therefore, cIMPACT-NOW (the Consortium to Inform Molecular and Practical Approaches to CNS Tumor Taxonomy) convened a working committee and released cIMPACT-NOW update 3, 5, and 6Jeny (30-32). According to cIMPACT-NOW update 3, IDH wild-type DA patients with EGFR amplification, or combined whole chromosome 7 gain and whole chromosome 10 loss, or TERT promoter mutation, should be considered WHO grade IV GBM (30). In this study, 6 patients with the IDH wild-type astrocytoma were found, and one of them also had a TERT promoter mutation. However, due to the lack of corresponding treatment guidelines for the patients with IDH wild-type astrocytoma, CNS WHO (2016) was still used as the main reference for diagnosis and classification in this study.
The KRT18 gene encodes type I intermediate filament chain keratin 18, which can provide the support for the cytoskeleton (33). The full-length fragment of KRT18 has been proposed to be used as a biomarker of liver damage (34). However, its function in glioma is still unknown. In this study, we found that somatic mutation of the KRT18 gene was significantly enriched in astrocytoma, but it was not found in GBM (42% vs. 0%). Furthermore, we found that all mutation sites of KRT18 in astrocytoma were located in the Coil 1A domain, which is the core region for the interaction of KRT18 with the TRADD and pinin proteins. TRADD is a tumor suppressor whose overexpression can lead to TNF-induced apoptosis and activation of nuclear factor (NF)-kappa-B (35). Moreover, it is also a direct regulator of cellular homeostasis, whose targeting may represent a promising strategy for restoring homeostasis to treat human diseases (17). Pinin is a nuclear and cell adhesion-related phosphoprotein, and its reduction can induce loss of cell-cell adhesion (36). It can bind to the transcriptional corepressor C-terminal binding protein (CtBP) and relieve CtBP-mediated repression of E-cadherin (18). Our results in this study indicate that the somatic mutation of KRT18 may affect its interaction with the TRADD protein and pinin protein, and change the apoptosis and adhesion of glioma cells. However, this notion needs to be verified by further experimentation.
CNV is another important molecular feature of glioma. In this study, we found that loss of MTAP was the most significant difference between astrocytoma and GBM. The MTAP gene is located at 9p21.3, which encodes 5'-methylthioadenosine phosphorylase, and plays a major role in polyamine metabolism (37). MTAP deletion frequently occurs in tumors, but its biological role in glioma remains unclear. Our analysis of the TCGA-GBM data suggested that loss of MTAP was associated with the poor prognosis of GBM. A study reported that loss of MTAP immunoreactivity can be used as a biomarker to predict the poor prognosis of glioma (38). MTAP loss could promote the stemness of GBM and confer a unique susceptibility to purine starvation (39). These studies indicated that MTAP may be a tumor suppressor gene in glioma. However, the pathologic consequence of MTAP loss in GBM remains unclear and need to further study.
ICIs have been approved for the treatment of multiple solid tumors. However, due to the inherent obstacles, such as the blood-brain barrier, tumor microenvironment, corticosteroids, and tumor heterogeneity, ICIs have limited therapeutic benefits for patients with glioma. An effective biomarker may improve the response of patients to ICIs. However, we found that, except for a few patients, it has a very low level of tumor mutational burden (TMB), microsatellite instability (MSI), and programmed death-ligand 1 (PD-L1) in glioma. We analyzed the infiltration of immune cells in the astrocytoma and GBM patients using the CIBERSORT Algorithm but found almost no obvious infiltration of CD8+T cells, B cells, CD4+T cells, or M1 macrophages. This indicated an immunosuppressive microenvironment. M2 macrophages were enriched in the TME of patients, but no significant difference was found between astrocytoma and GBM. A study reported that transforming growth factor beta 1 (TGF-β1) secreted by M2 macrophages could enhance the stemness and migration of glioma cells through the SMAD2/3 signaling pathway (40). Another study found that glioma patients with lower expression of the M2 marker, CD163, and higher expression of the M1 marker, CCL3, had better survival (41). These studies suggest that M2 macrophages are likely to be a common immune cell between astrocytoma and GBM, and closely related to tumorigenesis.
In addition, we also found a significant enrichment of M0 macrophages in GBM when compared with astrocytoma. Studies have shown that high-risk glioma populations usually have enrichment of M0 macrophages (42), and the infiltration of M0 macrophages is related to the poor prognosis of glioma (43). This is consistent with the poor prognosis of GBM. In the astrocytoma patients, we found significant enrichment of activated mast cells, monocytes, and plasma cells. Mast cells play an important role in allergies and allergic reactions (44). They can suppress the proliferation, migration, and stemness of glioma by downregulating the expression of GSK3β and inhibiting STAT3 activation (45). Monocytes are mononuclear myeloid cells that develop in bone marrow and circulate in the blood (46). The infiltration of monocytes has been shown to be positively related to prognosis in patients with glioma (43), and tends to be exhausted in patients with recurrent GBM (47). These studies suggest that the infiltration of immune cells in astrocytoma may be associated with its better prognosis compared to GBM.
In summary, we found that the somatic mutation of KRT18, which is associated with cell apoptosis and adhesion through interaction with TRADD and pinin, was significantly enriched in astrocytoma, but rare in GBM. Copy number loss of MTAP, which is closely related to the poor prognosis of glioma, was found to be significantly enriched in GBM. In addition, different SCNA and gene expression patterns between astrocytoma and GBM were also found. Furthermore, immune cell infiltration analysis showed that M0 macrophages were significantly enriched in GBM, while activated mast cells, monocytes, and plasma cells were significantly enriched in astrocytoma. This study revealed the distinct characteristics of astrocytoma and GBM at the DNA and RNA level, and may have an important value for exploring the occurrence of glioma.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-1317
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-1317
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-1317). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures involving human participants were in accordance with principles of the Declaration of Helsinki (2013). All patients signed informed consent, and the study design was approved by the Ethics Committee of Nanfang Hospital (NFEC-2019-006).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ostrom QT, Cioffi G, Gittleman H, et al. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2012-2016. Neuro-Oncol 2019;21:v1-100. [Crossref] [PubMed]
- Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 2016;131:803-20. [Crossref] [PubMed]
- Ghosh D, Nandi S, Bhattacharjee S. Combination therapy to checkmate Glioblastoma: clinical challenges and advances. Clin Transl Med 2018;7:33. [Crossref] [PubMed]
- Azzarelli R. Organoid Models of Glioblastoma to Study Brain Tumor Stem Cells. Front Cell Dev Biol 2020;8:220. [Crossref] [PubMed]
- van den Bent MJ, Smits M, Kros JM, et al. Diffuse Infiltrating Oligodendroglioma and Astrocytoma. J Clin Oncol 2017;35:2394-401. [Crossref] [PubMed]
- Ohgaki H, Kleihues P. The definition of primary and secondary glioblastoma. Clin Cancer Res 2013;19:764-72. [Crossref] [PubMed]
- Ajlan A, Recht L. Supratentorial low-grade diffuse astrocytoma: medical management. Semin Oncol 2014;41:446-57. [Crossref] [PubMed]
- Grimm SA, Chamberlain MC. Anaplastic astrocytoma. CNS Oncol 2016;5:145-57. [Crossref] [PubMed]
- Hartmann C, Hentschel B, Wick W, et al. Patients with IDH1 wild type anaplastic astrocytomas exhibit worse prognosis than IDH1-mutated glioblastomas, and IDH1 mutation status accounts for the unfavorable prognostic effect of higher age: implications for classification of gliomas. Acta Neuropathol 2010;120:707-18. [Crossref] [PubMed]
- Weller M, van den Bent M, Tonn JC, et al. European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol 2017;18:e315-29. [Crossref] [PubMed]
- Cieslik M, Chugh R, Wu YM, et al. The use of exome capture RNA-seq for highly degraded RNA with application to clinical cancer sequencing. Genome Res 2015;25:1372-81. [Crossref] [PubMed]
- Karn T, Meissner T, Weber KE, et al. A Small Hypoxia Signature Predicted pCR Response to Bevacizumab in the Neoadjuvant GeparQuinto Breast Cancer Trial. Clin Cancer Res 2020;26:1896-904. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]
- Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008;321:1807-12. [Crossref] [PubMed]
- Inada H, Izawa I, Nishizawa M, et al. Keratin attenuates tumor necrosis factor-induced cytotoxicity through association with TRADD. J Cell Biol 2001;155:415-26. [Crossref] [PubMed]
- Shi J, Sugrue SP. Dissection of protein linkage between keratins and pinin, a protein with dual location at desmosome-intermediate filament complex and in the nucleus. J Biol Chem 2000;275:14910-5. [Crossref] [PubMed]
- Xu D, Zhao H, Jin M, et al. Modulating TRADD to restore cellular homeostasis and inhibit apoptosis. Nature 2020;587:133-8. [Crossref] [PubMed]
- Alpatov R, Munguba GC, Caton P, et al. Nuclear Speckle-Associated Protein Pnn/DRS Binds to the Transcriptional Corepressor CtBP and Relieves CtBP- Mediated Repression of the E-Cadherin Gene. Mol Cell Biol 2004;24:10223-35. [Crossref] [PubMed]
- Feng J, Kim ST, Liu W, et al. An integrated analysis of germline and somatic, genetic and epigenetic alterations at 9p21.3 in glioblastoma. Cancer 2012;118:232-40. [Crossref] [PubMed]
- Honda T, Kobayashi K, Mikoshiba K, et al. Regulation of cortical neuron migration by the Reelin signaling pathway. Neurochem Res 2011;36:1270-9. [Crossref] [PubMed]
- Schulze M, Violonchi C, Swoboda S, et al. RELN signaling modulates glioblastoma growth and substrate-dependent migration. Brain Pathol 2018;28:695-709. [Crossref] [PubMed]
- Li X, Fan W, Yao A, et al. Downregulation of reelin predicts poor prognosis for glioma. Biomark Med 2020;14:651-63. [Crossref] [PubMed]
- Dao Trong P, Rösch S, Mairbäurl H, et al. Identification of a Prognostic Hypoxia-Associated Gene Set in IDH-Mutant Glioma. Int J Mol Sci 2018;19:2903. [Crossref] [PubMed]
- Korbecki J, Olbromski M, Dzięgiel P. CCL18 in the Progression of Cancer. Int J Mol Sci 2020;21:7955. [Crossref] [PubMed]
- Chang CY, Lee YH, Leu SJ, et al. CC-chemokine ligand 18/pulmonary activation-regulated chemokine expression in the CNS with special reference to traumatic brain injuries and neoplastic disorders. Neuroscience 2010;165:1233-43. [Crossref] [PubMed]
- Ma L, Wang H, Li Z, et al. Chemokine (C-C motif) ligand 18 is highly expressed in glioma tissues and promotes invasion of glioblastoma cells. J Cancer Res Ther 2019;15:358-64. [PubMed]
- Cheng W, Ren X, Zhang C, et al. Bioinformatic profiling identifies an immune-related risk signature for glioblastoma. Neurology 2016;86:2226-34. [Crossref] [PubMed]
- Weller M, Weber RG, Willscher E, Riehmer V, Hentschel B, Kreuz M, et al. Molecular classification of diffuse cerebral WHO grade II/III gliomas using genome- and transcriptome-wide profiling improves stratification of prognostically distinct patient groups. Acta Neuropathol 2015;129:679-93. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med 2015;372:2481-98. [Crossref] [PubMed]
- Brat DJ, Aldape K, Colman H, et al. cIMPACT-NOW update 3: recommended diagnostic criteria for ‘Diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV’. Acta Neuropathol 2018;136:805-10. [Crossref] [PubMed]
- Brat DJ, Aldape K, Colman H, et al. cIMPACT-NOW update 5: recommended grading criteria and terminologies for IDH-mutant astrocytomas. Acta Neuropathol 2020;139:603-8. [Crossref] [PubMed]
- Louis DN, Wesseling P, Aldape K, et al. cIMPACT-NOW update 6: new entity and diagnostic principle recommendations of the cIMPACT-Utrecht meeting on future CNS tumor classification and grading. Brain Pathol 2020;30:844-56. [Crossref] [PubMed]
- Wang PB, Chen Y, Ding GR, et al. Keratin 18 induces proliferation, migration, and invasion in gastric cancer via the MAPK signalling pathway. Clin Exp Pharmacol Physiol 2020; Epub ahead of print. [Crossref] [PubMed]
- Dear JW, Clarke JI, Francis B, et al. Risk stratification after paracetamol overdose using mechanistic biomarkers: results from two prospective cohort studies. Lancet Gastroenterol Hepatol 2018;3:104-13. [Crossref] [PubMed]
- Hsu H, Xiong J, Goeddel DV. The TNF receptor 1-associated protein TRADD signals cell death and NF-kappa B activation. Cell 1995;81:495-504. [Crossref] [PubMed]
- Joo JH, Alpatov R, Munguba GC, et al. Reduction of Pnn by RNAi induces loss of cell-cell adhesion between human corneal epithelial cells. Mol Vis 2005;11:133-42. [PubMed]
- Zhang T, Bauer C, Newman AC, et al. Polyamine pathway activity promotes cysteine essentiality in cancer cells. Nat Metab 2020;2:1062-76. [Crossref] [PubMed]
- Satomi K, Ohno M, Matsushita Y, et al. Utility of methylthioadenosine phosphorylase immunohistochemical deficiency as a surrogate for CDKN2A homozygous deletion in the assessment of adult-type infiltrating astrocytoma. Mod Pathol 2021;34:688-700. [Crossref] [PubMed]
- Hansen LJ, Sun R, Yang R, et al. MTAP Loss Promotes Stemness in Glioblastoma and Confers Unique Susceptibility to Purine Starvation. Cancer Res 2019;79:3383-94. [Crossref] [PubMed]
- Liu Z, Kuang W, Zhou Q, et al. TGF-β1 secreted by M2 phenotype macrophages enhances the stemness and migration of glioma cells via the SMAD2/3 signalling pathway. Int J Mol Med 2018;42:3395-403. [Crossref] [PubMed]
- Vidyarthi A, Agnihotri T, Khan N, et al. Predominance of M2 macrophages in gliomas leads to the suppression of local and systemic immunity. Cancer Immunol Immunother 2019;68:1995-2004. [Crossref] [PubMed]
- Zhu C, Zou C, Guan G, et al. Development and validation of an interferon signature predicting prognosis and treatment response for glioblastoma. Oncoimmunology 2019;8:e1621677 [Crossref] [PubMed]
- Zhong QY, Fan EX, Feng GY, et al. A gene expression-based study on immune cell subtypes and glioma prognosis. BMC Cancer 2019;19:1116. [Crossref] [PubMed]
- Nelissen S, Lemmens E, Geurts N, et al. The role of mast cells in neuroinflammation. Acta Neuropathol 2013;125:637-50. [Crossref] [PubMed]
- Attarha S, Roy A, Westermark B, et al. Mast cells modulate proliferation, migration and stemness of glioma cells through downregulation of GSK3β expression and inhibition of STAT3 activation. Cell Signal 2017;37:81-92. [Crossref] [PubMed]
- Mitchell AJ, Roediger B, Weninger W. Monocyte homeostasis and the plasticity of inflammatory monocytes. Cell Immunol 2014;291:22-31. [Crossref] [PubMed]
- Wang Q, Hu B, Hu X, et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell 2017;32:42-56.e6. [Crossref] [PubMed]


