
Hsa_circ_0080229 upregulates the expression of murine double minute-2 (MDM2) and promotes glioma tumorigenesis and invasion via the miR-1827 sponging mechanism
Introduction
Malignant glioma, which is the most frequent primary cranial tumor in adults, is known for its aggressive oncogenesis and deadly prognosis (1). The current standard therapy for gliomas is maximal safe surgical resection, followed by radiation therapy and temozolomide (TMZ) chemotherapy (2-4). However, as most patients with malignant gliomas still succumb to the disease within 2 years of diagnosis, there is an urgent demand for more effective therapeutic strategies (4).
Circular ribonucleic acids (circRNAs) are a new class of non-coding ribonucleic acids (RNAs) with a closed circular structure and are currently emerging as important players in cancer progression, malignancy, and metastasis (5,6). Recent studies have shown that circRNAs mainly affect competitive endogenous RNA (ceRNA) or micro-RNA (miRNA) sponges. Compared to linear long non-coding RNAs (lncRNA), circRNAs are more stable and effective at absorbing miRNAs, which are believed to target the 3' untranslated region (UTR) of downstream genes and subsequently decrease their expression (6,7). Some studies have shown that circRNAs are highly enriched in neuronal tissues, and their expressions are often regulated during neuronal development or tumorigenesis (8-11). Consequently, circRNAs likely mediate oncogenes or cancer suppressor genes by sponging miRNAs in gliomas. Li et al. reported that hsa_circ_0046701 promotes carcinogenesis by increasing Integrin Subunit Beta 8 by targeting miR-142-3p (9). Yang et al. found that a circRNA from F-box and WD-40 domain protein 7 plays a role in repressing glioma tumorigenesis (11).
The epidermal growth factor receptor (EGFR) is a prominent biomarker in many malignant cancers, including glioma (12-16). The amplification and overexpression of the EGFR gene have been reported to be a hallmark of primary glioblastoma, and a series of therapeutic methods targeting the EGFR have been applied in clinical trials (14,17). Such research has prompted us and many others to attempt to elucidate the mechanism related to EGFR.
Salzman et al. identified many circRNAs and the related linear transcripts for each gene and notably reported that hsa_circ_0080229 is a circRNA of the EGFR (18). To date, no research has been conducted to determine the role of circ_0080229 in gliomas. Consequently, their mechanism in tumorigenesis and progression remains unknown. We hypothesized that the circRNAs derived from the EGFR gene might be involved in oncogenesis.
In this study, we sought to identify the overexpression of circ_0080229 in gliomas and evaluate the association between circ_0080229 and gliomas classification based on the World Health Organization’s (WHO’s) grading standard (19). The function of circ_0080229 was verified in vitro and in vivo. Further, we tried to excavate the possible mechanism by which circ_0080229 mediates proliferation and invasion in gliomas.
We present the following article following the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-7123).
Methods
Tissue sample collection and cell cultures
Nineteen paired glioma tissues and adjacent normal tissues were obtained from patients who underwent surgery at Sun Yat-Sen Memorial Hospital from June 2016 to September 2017. All specimens were histopathologically confirmed and graded according to WHO’s staging guidelines. All samples from the surgery operations were immediately frozen and then stored in liquid nitrogen until RNA or protein extraction. The study was approved by the Drug Clinical Trial Institution Ethics Committee of Sun Yat-Sen Memorial Hospital (IRB number: 2016-161). The study conformed to the Declaration of Helsinki (as revised in 2013), and all patients provided their written and informed consent for all surgical treatments and molecular studies. Normal human astrocytes, human embryonic kidney (HEB) 293T and glioma U87 and U251 cell lines were purchased from the Chinese Academy of Sciences (Shanghai, China) and cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibico BRL) supplemented with 10% fetal bovine serum (FBS) (Gibico, qualified, South America).
Cell-line transfection and group division
RNAs (GenePharma, Shanghai) were transfected into the U87 and U251 cell lines using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). The following 3 main groups were used in this study: (I) a negative control (NC) group; (II) a si-circ_0080229 group; (III) a si+miR1827-inhibitor group. The oligos sequences of the NC, NC-inhibitor, and miR-1827 inhibitor were designed by GenePharma, Shanghai, China. The sense of si-circ_0080229 was designed as 5'-ACUGCACCUACGGUGAGCCTT-3', and the antisense as 5'-GGCUCACC GUAGGUGCAGUTT-3'. The transfection efficiency was confirmed by a quantitative real-time polymerase chain reaction (qRT-PCR) analysis.
RNA extraction and the qRT-PCR analysis
Total RNA was isolated from the cell lines or tissues using the TRIzol reagent (Invitrogen, USA) following the manufacturer’s instructions. We then transferred RNA to complementary deoxyribonucleic acid (cDNA) using the PrimeScript™ Reverse Transcription Kit (TakaRa, Japan). To prepare the miRNA’s cDNA, we followed the instructions of the Mir-X miRNA First-Strand Synthesis Kit (TakaRa, Japan). The primers were designed as described in Table 1 and synthesized by Guangzhou IGE Biotechnology LTD (Guangzhou, China). We then performed a qRT-PCR analysis using a FastStart Universal SYBR-Green Master (TakaRa Japan). The following procedures were performed in PCR: (I) the activation of enzymes at 95 °C for 5 minutes; (II) the denaturation at 95 °C for 20 seconds; (III) the annealing and extension at 60 °C for 30 seconds; (IV) the run 45 cycles for (II) and (III). The relative expression levels of the PCR products were calculated using the 2−ΔΔCt method. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or vampyrus U6 spliceosomal RNA (U6) were used as the internal references.
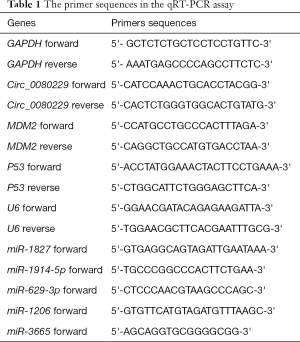
Full table
In-situ hybridization (ISH) and fluorescence in-situ hybridization (FISH) testing
The probe sequence of the ISH kit was “TCCCTTGGCTCACCGTAGTGCAG”. The probe was designed to target the circular conjunction of circ_0080229, which made it specific to the circRNAs rather than its linear transcripts. First, we prepared paraffin-embedded slices that were dewaxed and rehydrated according to standard procedures. Next, we used 3% hydrogen peroxide (H2O2) to treat the slices for 10 minutes to inactivate endogenous enzymes. Proteinase K (20 µg/mL) was applied to the slices to expose nucleic acid segments at 37 °C for 30 minutes. We dropped the probe-based hybridization solution onto the slices at 73 °C for 5 minutes, put the slices into pre-hybridization at 37 °C for 2 hours, and further incubated them at 37 °C for 16–18 hours. The blocking solution was dropped onto the slices at 37 °C for 30 minutes. Biotinylated mouse anti-digoxin was added to the slices for the reaction at 37 °C for 1 hour. After being washed 3 times with PBS, streptavidin-biotin complex peroxidase was then dropped to react at 37 °C for 30 minutes before washing with PBS. Finally, diaminobenzidine and hematoxylin were added to the slides.
The procedures for the FISH assay were similar to those of the ISH assay except that the FISH probe was attached with cyanine 3, which was detected with a fluorescent microscope. Both the ISH and FISH Kits and designed probes were purchased from Ribobio (Guangzhou, China).
Western blot analysis
Proteins from cell lines or tissues were pyrolyzed using a Ristocetin-Induced Platelet Aggregation Lysis Buffer (Beyotime, China), and then denatured at 98 °C for 10 minutes. The total proteins in lysates were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to a 0.22-µm polyvinylidene difluoride membrane (Millipore, Billerica, MA, USA). The membrane was subsequently incubated with 5% bovine albumin, and specific antibodies were applied to incubate the proteins. Finally, blotting was imaged following the manufacturer’s protocol. The GAPDH antibody was the internal preference. The primary antibodies were as follows: GAPDH (Abcam, ab8245, Cambridge, MA, USA); MDM2 (Abcam, ab16895, Cambridge, MA, USA); and P53 (Abcam, ab26, Cambridge, MA, USA).
Dual-luciferase reporter gene detection assay
The luciferase reporter plasmid was constructed by BersinBio (Guangzhou, China), and the targeted sequence (wild type) of MDM2 or circ_0080229 was added to the downstream of Firefly Luciferase; Renilla Luciferase was used as an internal reference. The mutant-type reporter was also used as a model control group to eliminate any irrelevant plasmid interference.
Next, 293T cells were cultured in 24-well plates, which were co-transfected with the reporter plasmid and miR-1827 mimics or an inhibitor or the NC oligos. After co-incubation for about 48 hours, the cells were extracted, and the relative intensity of the Firefly luciferase (compared to Renilla Luciferase) was examined in accordance with the manufacturer’s protocol (Yeasen, Shanghai, China).
Wound-healing assay
After general cell culture, cells were seeded into 6-well plates with a density of 1×106/well. The cells were put into incubation and cultured to 90% confluence in about 24–48 hours. A sterile pipette tip of 100 µL was used to scratch the cell layers to form wounded gaps. The plates were gently washed with PBS, and DMEM (without FBS) was then added to the plates that were incubated at 37 °C for 48 hours. The wound gaps were photographed with a microscope.
Transwell migration and invasion assay
The invasion assay needed transwell inserts (8.0 µm, Costar, USA) were coated with 50 µL of 1 mg/mL Matrigel matrix (BD, Franklin, USA) following the manufacturer’s guidelines. U87 or U251 cells (8×104) in 200 µL DMEM (without FBS) were plated in the upper chamber, and 500 µL cultured media with FBS were added to the lower chamber. The migration assay was similar to the invasion assay, except that the transwell insert was not coated with Matrigel. After incubation for 24 hours at 37 °C in a humid atmosphere with 5% carbon dioxide (CO2), the cells that did not penetrate the membrane were gently removed by a swab, while the migrated or invaded cells were fixed with 4% paraformaldehyde, stained with 0.1% crystal violet and photographed with a microscope.
3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) assay
Briefly, U87 or U251 cells (5×103) in different groups were seeded into 96-well plates and cultured at 37 °C in a humid atmosphere with 5% CO2. After 72 hours of incubation, the cells were treated with MTT solution (1 mg/mL) for 3 hours. Then, the culture medium was removed, and dimethyl sulfoxide (100 µL) was added to each well for 30 minutes of incubation. Optical density (OD) values were detected with a Multiscan Spectrum (Thermo, MA, USA) to reflect proliferation.
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymetho-xyphenyl)-2-(4-sulfopheny)-2H-tetrazolium inner salter (MTS) assay
Similar to the MTT assay, an MTS assay was applied to detect proliferation. U87 or U251 cells (5×103) in the 3 groups (see above) were seeded into 96-cell plates and cultured at 37 °C with 5% CO2. After certain time points (24, 48, 72, and 96 hours), the cells were treated with MTS (KeyGEN, China) for 1 hour, and the OD values were detected to represent the proliferation following the manufacturer’s protocol.
Colony formation assay
U87 and U251 cells (500/well) in different groups were seeded into 6-well plates and cultured for 10 days. The colonies were then fixed with 4% paraformaldehyde for 20 minutes and stained with 0.1% crystal violet for 10 minutes. The number of colonies was counted, and the colony formation rate was calculated.
EdU (5-ethynyl-2'-deoxyuridine) cell proliferation assay
U87 and U251 cells cultured in 6-well plates were treated with EdU solution (10 µM) for 2 hours. The cells were then treated with 4% paraformaldehyde for 30 minutes, and the plates were washed with PBS 3 times. Finally, the EdU-FITC fluorescence was detected using the EdU Cell Proliferation Kit with Alexa Fluor 488 (BeyoClick, China) following the manufacturer’s instructions.
Annexin V-fluorescein isothiocyanate (FITC)/polyimides (PI) apoptosis assay
For the cell apoptosis analysis, U87 and U251 cells were first treated with small interfering RNA (siRNA) of circ_0080229 and miR-1827 and divided into the 3 groups (see above). Then, TMZ was used to co-induce apoptosis in different groups of cells. After 48 hours, the cells were collected. The cells (5×105) were stained successively with Annexin V-FITC and PI, and flow cytometry was used to detect the fluorescence, which reflected the intensity of Annexin V or PI.
TdT-mediated dUTP nick end labeling (TUNEL) assay
An In-Situ Cell Death Detection Kit, POD (Roche, CH, Switzerland) was used to assess the apoptosis based on TUNEL technology following the manufacturer’s protocol. First, U87 and U251 cells were induced with TMZ for 48 hours and then were fixed by 4% triformol paraformaldehyde for 30 minutes. Then, 3% H2O2 was dropped onto the samples for permeabilization before the TUNEL reaction mixture was added. Finally, after incubation with the TUNEL reaction mixture, the samples were analyzed by fluorescence microscopy.
In-vivo tumorigenesis assay
BALB/c nude mice (aged 4 weeks; half male and female) were purchased from Beijing Vital River Laboratory Co. Ltd. (China) and maintained in specific pathogen-free conditions at the Animal Experiment Centre, Sun Yat-Sen University. Twelve mice were randomly divided into the following 2 groups using a simple random sampling method: (I) an NC group; (II) a si-circ_0080229 group. U251 cells were first transfected with either si-circ_0080229 oligo or the NC oligo, and 5×106 cells were then subcutaneously injected into the right flank of each nude mouse. The weight of the mice and the size of the tumors were monitored and recorded every 5 days, and the tumor volume was estimated using the following formula: V = ab2/2 (where “a” was the variable for the major axis and “b” for the minor axis). When the average tumor volume was about 13.5 mm3, the NC or the si-circ_0080229 RNA oligo was injected into the tumors 5 times every 7 days. At the end of the experiment, the mice were put to death, and the tumors’ weights were recorded. Further, each tumor was stained with hematoxylin and eosin to verify its pathology. All animal experiments were approved by the Animal Experiment Ethics Committee of Sun Yat-Sen University (LACUC number: L102022016000D) and conducted following the National Research Council (USA) guidelines on the care and use of laboratory animals (20).
Statistical analysis
The significance of the differences between the groups was estimated using the Student’s t-test, one-way analysis of variances, or Chi-square test as appropriate. A two-tailed P<0.05 was considered statistically significant. All of the statistical analyses were performed with SPSS 19.0 software (IBM, SPSS, Chicago, IL, USA).
Results
The upregulation of circ_0080229 is correlated with an unfavorable prognosis in gliomas
To identify the expression of circ_0080229 in gliomas, ISH testing and qRT-PCR analysis were conducted in samples of patients with gliomas and pair-matched adjacent normal tissues. Figure 1A shows the ISH of circ_0080229 expression. In Figure 1A,B,C,D, circ_0080229 is shown to be significantly upregulated in glioma tissues, and the higher-grade glioma tissues show more abundant staining than the lower-grade glioma tissues. Circ_0080229 was mainly localized in the cytoplasm of the tissue cells (see Figure 1A). The ISH samples’ quantification analysis indicated glioma grade differential upregulation, which suggests that circ_0080229 expression is correlated with malignancy (see Figure 1B). To further verify the association between the circ_0080229 expression and the glioma WHO grade, FISH testing was conducted on a tissue chip of 80 samples with different WHO grades. The results showed that high circ_0080229 expression was related to high-grade gliomas, as shown in the heatmap (see Figure 1C). Quantification by a qRT-PCR analysis further proved the upregulation of circ_0080229 expression in gliomas (see Figure 1D). The expression of circ_0080229 was detected in different cell lines, and the results showed that the expression of circ_0080229 in the U87 and U251 cell lines was significantly increased compared to that in HEB cell lines (see Figure 1E). Moreover, to investigate the clinical significance of circ_0080229, we analyzed its expression level and matched clinicopathological characteristics and found out that circ_0080229 expression level was significantly higher in patients at an advanced stage or with advanced tumor sizes (see Table 2).
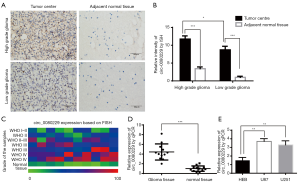
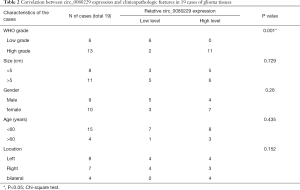
Full table
circ_0080229 knockdown inhibited the proliferation of glioma cells
Having confirmed the high expression of circ_0080229 in the U87 and U251 glioma cell lines, these cell lines were used to elucidate the importance of circ_0080229 in the migration and invasion of glioma. We proceeded to block circ_0080229 using siRNA to investigate the role of circ_0080229 in the migration and invasion of glioma cells. Next, to determine the rescue effect of miR-1827, the cells were co-treated with si-circ_0080229 and the mi-1827 inhibitor. The cells were then divided into the following 3 groups: (I) non-treated control (NC); (II) si-circ_0080229 treated cells; and (III) si-circ_0080229 + mi-1827 inhibitor-treated cells.
The colony formation assay showed a reduction in the colony formation rate in the si-circ_0080229 group in both the U87 and U251 cell lines compared to the NC group and was restored to some extent in the si+miR1827 inhibitor group (see Figure 2A,B). The OD values in the MTS assay demonstrated the growth trend in the 3 groups in the U87 or U251 cell lines, which indicated that the knockdown of circ_0080229 moderated the rising trend, while also inhibiting the expression of miR-1827, which in turn led to some improvement (see Figure 2C,D). Conversely, we sought to confirm MDM2 as the downstream target of the circ_0080229-mediated mechanism using a MTT assay. The results showed that the knockdown of circ_0080229 decreased proliferation while the overexpression of MDM2 significantly rescued the trend in the U87 and U251 cell lines (see Figure 2E,F). Meanwhile, the EdU (5-ethynyl-2'-deoxyuridine) proliferation assay showed a significant decrease in the proliferation rate in the si-circ_0080229 group, a result which was reversed in the si+miR-1827 inhibitor group (see Figure 2G,H).
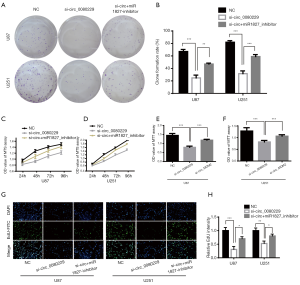
circ_0080229 knockdown suppressed the migration and invasion of glioma cells
Compared to the NC and si+miR1827-inhibitor group, the migration ability proven by the wound healing assay was significantly attenuated after 48 hours in the si-circ_0080229 treated group in both the U87 and U251 cell lines (see Figure 3A,B). Additionally, the transwell migration assay showed a significant decline in migration ability when it was treated with si-circ_0080229; however, this was reversed with the miR-1827 inhibitor (see Figure 3C,D). Further, the transwell invasion assay demonstrated that the invasive potential in group si-circ_0080229 was weaker than that of the NC group and that simultaneously transfecting cells with si-circ_0080229 and miR-1827 inhibitor subsequently reversed the invasive ability (see Figure 3E,F).
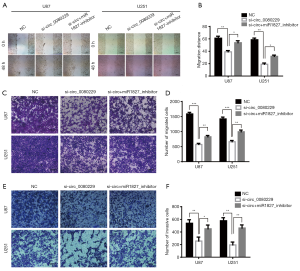
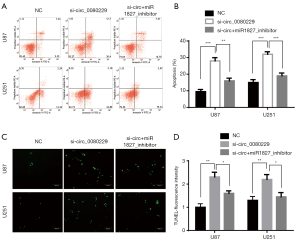
circ_0080229 knockdown enhanced the apoptosis induction in TMZ-treated gliomas cells
TMZ is the first-line treatment for glioblastoma, and it is commonly used in different kinds of research that seek to induce apoptosis of glioma. To determine the effect of circ_0080229 in apoptosis, U87 and U251 cells were treated with siRNA of circ_0080229 and miR-1827 and divided into 3 groups. Then, U87 cells in the different groups were induced with 200 µmol/L TMZ, and the U251 cells in all groups were treated with 320 µmol/L TMZ based on the general median lethal dose of each cell line. After 36 hours of induction with TMZ, a flow cytometry analysis was performed to detect apoptosis in both the U87 and U251 cell lines. The results indicated that the si-circ_0080229 group had a higher apoptosis rate than that of the NC or si+miR1827-inhibitor group (see Figure 4A,B). Similar results were found in the TUNEL analysis of the different groups; that is, that knockdown of circ_0080229 was found to enhance apoptosis induced by TMZ in the U251 and U87 cell lines, which could be reversed by inhibiting miR-1827 (see Figure 4C,D).
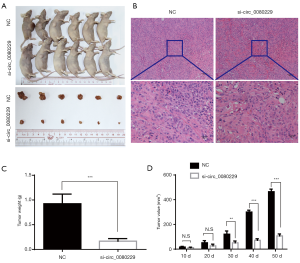
circ_0080229 knockdown inhibited tumor growth in vivo
To further explore whether circ_0080229 affected tumor growth in vivo, we constructed a xenograft subcutaneous tumor model in BALB/c nude mice and then treated the mice with either si-circ_0080229 or NC with GenEscort™ III. On average, the xenograft tumor volume took about 10 days to grow up to 13.5 mm3. After that, intra-tumoral injections were performed 5 times every 7 days with either si-circ_0080229 or NC, depending on the group. These treatments did not produce any side effects or adverse events in the mice. Sixty days after treatment, 6 mice in each group were sacrificed, and the subcutaneous tumors were isolated and examined (see Figure 5A). The pathology of the tumors was verified using hematoxylin and eosin staining (see Figure 5B). On average, the tumor weight was 0.167±0.090 g in the si-circ_0080229 group and 0.924±0.338 g in the NC group; thus, tumors in the si-circ_0080229 group were significantly lighter than those in the NC group (see Figure 5C). The tumor sizes were also detected, and the growth curve of tumors in the si-circ_0080229 group was significantly inhibited compared to that of the NC treated group (see Figure 5D).
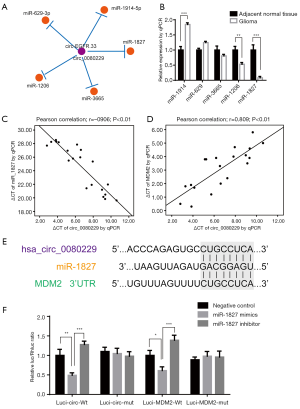
circ_0080229 physically binds to miR-1827 that targets the 3'UTR of MDM2
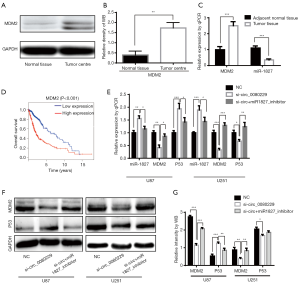
Using circBase (http://www.circbase.org), we confirmed that hsa_circ_0080229 was the same circRNA as hsa-circ-EGFR.33 on CircNet (http://circnet.mbc.nctu.edu.tw). The following top 5 microRNAs, which were the potential target of circ_0080229, were also identified via CircNet: miR-1827, miR-3665, miR-1206, miR-629-3p, and miR-1914-5p (see Figure 6A). A qRT-PCR analysis was performed to examine the expression of each of these 5 microRNAs in glioma tissues and adjacent normal tissues. Among all the miRNAs, miR-1827 had the most significant downregulation in glioma tissues than adjacent normal tissues (P<0.001; see Figure 6B). As miR-1827 is involved in the MDM2 pathway in colorectal cancer (21), we further analyzed the related expression of MDM2 in gliomas. A qRT-PCR analysis of the 20 glioma samples showed that circ_0080229 and miR-1827 were negatively correlated (P<0.01; Pearson correlation –0.906; see Figure 6C), while circ_0080229 and MDM2 were positively correlated (P<0.01; Pearson correlation 0.809; see Figure 6D). Notably, the bioinformatic analysis revealed that miR-1827 contained a putative binding site of circ_0080229 while simultaneously binding a 3'UTR of MDM2 (see Figure 6E). To verify the binding sites between circ_0080229 and miR-1827 and those between miR-1827 and MDM2, a dual-luciferase reporter gene expression assay was performed in the 293T cell line, which is frequently used in general luciferase reporter gene assays. The luciferase reporter expression intensity was significantly attenuated by miR-1827 mimics targeting the plasmid of both MDM2 and circ_0080229 wild-type sequence. However, there was no difference in the luciferase reporter assay results concerning the mutant control group with the MDM2 or circ_0080229 sequence. These results validated the binding potency between miR-1827 and circ_0080229 and between miR-1827 and the MDM2 3'UTR (see Figure 6F).
circ_0080229 regulated MDM2 expression by competitively sponging miR-1827
miR-1827 has been reported to act as a tumor suppressor targeting the 3'UTR of MDM2 and its downstream P53 pathway in colorectal cancer (21). Thus, we hypothesized that miR-1827 played a similar role in glioma and tried to illuminate further the signaling pathway of MDM2 and P53 induced by circ_0080229. The expressions of MDM2 were increased in glioma tissues compared to those of adjacent normal tissues, as illustrated by qRT-PCR analysis and a western blot analysis (see Figure 7A,B). Further, their expressions were negatively correlated with miR-1827 expression (see Figure 7C). A survival analysis based on The Cancer Genome Atlas (TCGA) showed that a high MDM2 level was correlated with an unfavorable prognosis in glioma (see Figure 7D). The knockdown of circ_0080229 in the U87 and U251 cell lines decreased MDM2 while miR-1827 inhibitor transfection rescued the reduction. Interestingly, circ_0080229 silencing led to the suppression of P53 in U87 and overexpression in U251(see Figure 7E,F,G). The above results indicated that circ_0080229 could sponge miR-1827 and result in the overexpression of the oncogene MDM2.
Discussion
Glioma is one of the deadliest cancers for which there is currently no efficient therapy. Due to its indistinct mechanism, the clinical treatment of glioma represents a great challenge. The EGFR has been shown to play an important role in the occurrence and treatment of glioma, but the detailed mechanism by which it does so remains unclear. Recent studies have shown that circRNAs play critical roles in the tumorigenesis and metastasis of most cancers, including glioma (5,6,22,23). Some studies have demonstrated the novel roles of circRNAs in glioma, such as Tau tubulin kinase 2 circRNA and hsa_circ_0046701 (9), whose main mechanisms are to mediate the downstream gene by sponging miRNAs. It was reasonable to deduce that the EGFR-derived circRNA might also be important in glioma. However, to date, no study had reported on the function of circ_0080229 in glioma.
In the present study, we identified circ_0080229 as a novel biomarker in glioma. Circ_0080229 is upregulated in glioma cell lines and tissues and is associated with the clinicopathologic stage. Functional assays of circ_0080229 in the U87 and U251 glioma cell lines indicated that circ_0080229 knockdown suppressed the proliferation, migration, and invasion of glioma and also enhanced the induction of apoptosis in TMZ-treated cells. Also, the treatment of si-circ_0080229 in a subcutaneous xenograft tumor model in BALB/c nude mice indicated that circ_0080229 promoted tumor growth and progression in vivo. Concerning the mechanism, circ_0080229 could sponge miR-1827 while the latter targeted the 3'UTR of MDM2 to decrease MDM2 expression, which was shown by a dual-luciferase reporter, a qRT-PCR analysis, and a western blot analysis. In conclusion, circ_0080229 promoted glioma tumorigenesis and metastasis by increasing MDM2 expression by sponging miR-1827.
It has been clearly shown that MDM2 and wild-type P53 gene constitute a feedback-loop inside which MDM2 decreases the tumor suppressor P53 (24-26). However, in addition to mediating the P53 pathway, MDM2 also exerts its oncogene functions on many other signaling pathways (27-29); for example, it also interacts with the histone methyltransferase SUV39H1 (30,31). Thus, as reported (26), the expression of MDM2 is also increased in some P53-mutant tumors. In the present study, circ_0080229 was found to increase the expression of MDM2, which illustrated the regulatory mechanism of the circRNA derived from the EGFR.
As mentioned above, MDM2 always decreases the wild-type P53 gene. Our research found a notable and different trend in P53 expression after silencing circ_0080229 between U87 and U251, whereby the former was increased while the latter was decreased. This was because the P53 gene in U87 is a wild type, and U251 contains a mutant P53 gene (mutant at codon 273; CGT/CAT; Arg/His) (32). The wild-type P53 plays a critical role in tumor suppression, while the mutant P53 deteriorates into an oncogene (2,33,34). In a cell line with a wild-type P53, such as U87, circ_0080229 increases MDM2 to suppress the expression of the tumor suppressor P53. Conversely, in a cell line with mutant P53, such as U251, circ_0080229 increases the expression of the mutant P53, the oncogene in glioma.
MiRNAs have been widely reported to play a critical role in mediating gene expression by binding with the 3'UTR (35-38). Concerning miR-1827, it has been reported to be a novel regulator in colorectal carcinoma and small-cell lung cancer (21,39-41). Zhang et al. showed that microRNA-1827 suppressed tumorigenesis by repressing MDM2 expression in colorectal cancer (40). In the present study, we elucidated the function of miR-1827 as an MDM2 mediator by targeting the 3'UTR of MDM2 and further discovered that circ_0080229 was a regulator of miR-1827. Thus, miR-1827 served as a bridge between circ_0080229 and MDM2, indicating circ_0080229 mediates tumorigenesis and invasion in glioma by targeting the MDM2 pathway.
EGFR plays an important role in glioma; however, very few studies have examined circRNA derived from the EGFR. Our work creatively showed that the EGFR-derived circ_0080229 is a critical biomarker in glioma and further illuminated its regulatory mechanism by sponging miR-1827 to the target MDM2/P53 pathway (see Figure 8).
Conclusions
Taken together, all the analyses indicated that circ_0080229 is an oncogenic factor that mechanistically promotes proliferation and metastasis in glioma by increasing MDM2 by sponging miR-1827. Thus, the circRNA hsa_circ_0080229 has the potential to act as a diagnostic and prognostic biomarker in glioma.
Acknowledgments
Funding: This work was supported by a grant from the National Natural Science Foundation of China (award number: 81602205) and the Young Teacher Training Project of Sun Yat-Sen University (award number: 20ykpy97).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-7123
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-7123
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-7123). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All samples were approved by our institutions’ ethical committees and obtained from the Departments of Neurosurgery at our institutions (i.e., the Sun Yat-Sen Memorial Hospital and Sun Yat-Sen University, 2016-161). The study conformed to the Declaration of Helsinki (as revised in 2013), and all patients provided their written and informed consent for all surgical treatments and molecular studies. All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC number: L102022016000D), Sun Yat-Sen University, and were conducted following the National Research Council (US) Committee guidelines on the care and use of laboratory animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Alexander BM, Cloughesy TF. Adult Glioblastoma. J Clin Oncol 2017;35:2402-9. [Crossref] [PubMed]
- Appin CL, Brat DJ. Biomarker-driven diagnosis of diffuse gliomas. Mol Aspects Med 2015;45:87-96. [Crossref] [PubMed]
- Hu J, Western S, Kesari S. Brainstem Glioma in Adults. Front Oncol 2016;6:180. [Crossref] [PubMed]
- Gan HK, van den Bent M, Lassman AB, et al. Antibody-drug conjugates in glioblastoma therapy: the right drugs to the right cells. Nat Rev Clin Oncol 2017;14:695-707. [Crossref] [PubMed]
- Chen B, Huang S. Circular RNA: An emerging non--coding RNA as a regulator and biomarker in cancer. Cancer Lett 2018;418:41-50. [Crossref] [PubMed]
- He J, Xie Q, Xu H, et al. Circular RNAs and cancer. Cancer Lett 2017;396:138-44. [Crossref] [PubMed]
- Hsiao KY, Sun HS, Tsai SJ. Circular RNA - New member of non-coding RNA with novel functions. Exp Biol Med (Maywood) 2017;242:1136-41. [Crossref] [PubMed]
- Chen W, Schuman E. Circular RNAs in Brain and Other Tissues: A Functional Enigma. Trends Neurosci 2016;39:597-604. [Crossref] [PubMed]
- Li G, Yang H, Han K, et al. A novel circular RNA, hsa_circ_0046701, promotes carcinogenesis by increasing the expression of miR-142-3p target ITGB8 in glioma. Biochem Biophys Res Commun 2018;498:254-61. [Crossref] [PubMed]
- Song X, Zhang N, Han P, et al. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res 2016;44:e87 [Crossref] [PubMed]
- Yang Y, Gao X, Zhang M, et al. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J Natl Cancer Inst 2018;110:304-15. Erratum in: J Natl Cancer Inst 2018 Oct 1;110(10):1147. doi: 10.1093/jnci/djy118. [Crossref] [PubMed]
- Di Sapio A, Morra I, Pradotto L, et al. Molecular genetic changes in a series of neuroepithelial tumors of childhood. J Neurooncol 2002;59:117-22. [Crossref] [PubMed]
- Fulci G, Labuhn M, Maier D, et al. p53 gene mutation and ink4a-arf deletion appear to be two mutually exclusive events in human glioblastoma. Oncogene 2000;19:3816-22. [Crossref] [PubMed]
- Hegi ME, Rajakannu P, Weller M. Epidermal growth factor receptor: a re-emerging target in glioblastoma. Curr Opin Neurol 2012;25:774-9. [Crossref] [PubMed]
- Nagane M. Genetic alterations and biomarkers for glioma. Brain Nerve 2012;64:537-48. [PubMed]
- Komotar RJ, Starke RM, Connolly ES, et al. Alteration in NFKBIA and EGFR in glioblastoma multiforme. Neurosurgery 2011;68:N14-5.
- Osuka S, Van Meir EG. Overcoming therapeutic resistance in glioblastoma: the way forward. J Clin Invest 2017;127:415-26. [Crossref] [PubMed]
- Salzman J, Chen RE, Olsen MN, et al. Cell-type specific features of circular RNA expression. PLoS Genet 2013;9:e1003777 [Crossref] [PubMed]
- Aldape K, Zadeh G, Mansouri S, et al. Glioblastoma: pathology, molecular mechanisms and markers. Acta Neuropathol 2015;129:829-48. [Crossref] [PubMed]
- National Research Council Committee for the Update of the Guide for the C, Use of Laboratory A. The National Academies Collection: Reports funded by National Institutes of Health. Guide for the Care and Use of Laboratory Animals. Washington (DC): National Academies Press (US) Copyright © 2011, National Academy of Sciences, 2011.
- Xiong F, Wu C, Chang J, et al. Genetic variation in an miRNA-1827 binding site in MYCL1 alters susceptibility to small-cell lung cancer. Cancer Res 2011;71:5175-81. [Crossref] [PubMed]
- Yang P, Qiu Z, Jiang Y, et al. Silencing of cZNF292 circular RNA suppresses human glioma tube formation via the Wnt/β-catenin signaling pathway. Oncotarget 2016;7:63449-55. [Crossref] [PubMed]
- Xie H, Ren X, Xin S, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget 2016;7:26680-91. [Crossref] [PubMed]
- Lane DP, Cheok CF, Lain S. p53-based cancer therapy. Cold Spring Harb Perspect Biol 2010;2:a001222 [Crossref] [PubMed]
- Brown CJ, Lain S, Verma CS, et al. Awakening guardian angels: drugging the p53 pathway. Nat Rev Cancer 2009;9:862-73. [Crossref] [PubMed]
- Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer 2013;13:83-96. [Crossref] [PubMed]
- Shuvalov O, Kizenko A, Shakirova A, et al. Nutlin sensitizes lung carcinoma cells to interferon-alpha treatment in MDM2-dependent but p53-independent manner. Biochem Biophys Res Commun 2018;495:1233-9. [Crossref] [PubMed]
- Qin JJ, Li X, Wang W, et al. Targeting the NFAT1-MDM2-MDMX Network Inhibits the Proliferation and Invasion of Prostate Cancer Cells, Independent of p53 and Androgen. Front Pharmacol 2017;8:917. [Crossref] [PubMed]
- Dimitriadi M, Poulogiannis G, Liu L, et al. p53-independent mechanisms regulate the P2-MDM2 promoter in adult astrocytic tumours. Br J Cancer 2008;99:1144-52. [Crossref] [PubMed]
- Chen L, Li Z, Zwolinska AK, et al. MDM2 recruitment of lysine methyltransferases regulates p53 transcriptional output. Embo J 2010;29:2538-52. [Crossref] [PubMed]
- Mungamuri SK, Benson EK, Wang S, et al. p53-mediated heterochromatin reorganization regulates its cell fate decisions. Nat Struct Mol Biol 2012;19:478-84, S1.
- Kataoka Y, Murley JS, Patel R, et al. Cytoprotection by WR-1065, the active form of amifostine, is independent of p53 status in human malignant glioma cell lines. Int J Radiat Biol 2000;76:633-9. [Crossref] [PubMed]
- Pykett MJ, Azzam E, Dahlberg W, et al. Differential p53, p21, mdm2 and Rb regulation in glioma cell lines that overexpress wild-type p53. Int J Oncol 1998;13:213-6. [Crossref] [PubMed]
- Rubio MP, von Deimling A, Yandell DW, et al. Accumulation of wild type p53 protein in human astrocytomas. Cancer Res 1993;53:3465-7. [PubMed]
- Rizvi NF, Smith GF. RNA as a small molecule druggable target. Bioorg Med Chem Lett 2017;27:5083-8. [Crossref] [PubMed]
- Liu J, Zhang C, Zhao Y, et al. MicroRNA Control of p53. J Cell Biochem 2017;118:7-14. [Crossref] [PubMed]
- Cora' D, Re A, Caselle M, et al. MicroRNA-mediated regulatory circuits: outlook and perspectives. Phys Biol 2017;14:045001 [Crossref] [PubMed]
- Mikaelian I, Scicchitano M, Mendes O, et al. Frontiers in preclinical safety biomarkers: microRNAs and messenger RNAs. Toxicol Pathol 2013;41:18-31. [Crossref] [PubMed]
- Fasihi A, M Soltani B, Atashi A, et al. Introduction of hsa-miR-103a and hsa-miR-1827 and hsa-miR-137 as new regulators of Wnt signaling pathway and their relation to colorectal carcinoma. J Cell Biochem 2018;119:5104-17. [Crossref] [PubMed]
- Zhang C, Liu J, Tan C, et al. microRNA-1827 represses MDM2 to positively regulate tumor suppressor p53 and suppress tumorigenesis. Oncotarget 2016;7:8783-96. [Crossref] [PubMed]
- Zhu S, Peng W, Li X, et al. miR-1827 inhibits osteogenic differentiation by targeting IGF1 in MSMSCs. Sci Rep 2017;7:46136. [Crossref] [PubMed]



