
Transcriptome sequencing and analysis reveals the molecular mechanism of skeletal muscle atrophy induced by denervation
Introduction
Skeletal muscle is an important effector organ of the peripheral nervous system. The integrity of its structure and maintenance of its function is controlled and regulated by the peripheral nervous system. Peripheral nerve injury leads to the loss of innervation of skeletal muscle, and then the loss of signal transmission and material exchange with nerves, resulting in a series of pathological changes, such as the reduction of cross-sectional area of muscle fibers, destruction of myofilaments and sarcomeres, degradation of myofibrils, decrease of contraction speed and fibrosis, and finally leads to skeletal muscle atrophy (1-3). Due to the slow speed of nerve regeneration after peripheral nerve injury, severe cases often experience irreversible atrophy or even disability before the target muscle is reinnervated, which brings a heavy burden to their families and broader society (4). Therefore, in-depth study of the molecular mechanism of muscle atrophy caused by peripheral nerve injury and exploration of new effective drug targets for prevention and treatment of skeletal muscle atrophy are important links to solve a series of problems including targeted muscle repair and functional reconstruction after peripheral nerve injury.
The characteristics of muscle atrophy are decreased muscle mass, activation of protein hydrolysis pathway, and inhibition of protein synthesis pathway activity. In particular, activation of the proteolytic pathway is an important cause of virtually all muscle atrophy (5). The proteolytic systems mainly include ubiquitin proteasome system (UPS), autophagy lysosome system (ALS), cathepsin hydrolysis system, and so on. The activation of these proteolytic pathways in the process of skeletal muscle atrophy involves a very complex molecular mechanism (6). Following activation, the proteolytic pathways hydrolyze myofibrils and cause skeletal muscle atrophy. During muscle atrophy, a series of biochemical and physiological events occurs in the atrophying muscles, which are closely related to the expression of many genes in muscles. Previous studies have examined many differentially expressed genes (DEGs) in atrophying muscles by using complementary DNA (cDNA) microarray (5,7-10). Several molecular mediators in skeletal muscles have been identified to play crucial roles in denervation-induced muscle atrophy (11-18). However, it is still unclear which biological processes are affected in the process of denervation-induced muscle atrophy. This is neither conducive to further exposing the molecular regulatory mechanism of denervation-induced muscle atrophy, nor to providing a potential target for the prevention and treatment of denervation-induced muscle atrophy.
Therefore, the present study aimed to describe the transcriptional landscape of rat tibialis anterior (TA) muscles after denervation. In this study, we constructed a model of denervation-induced muscle atrophy. Transcriptome sequencing technology was used to analyze the gene expression changes in denervated skeletal muscle and analyze the DEGs. Furthermore, cluster analysis, Gene Ontology (GO), and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were used to analyze the DEGs, which revealed some interesting aspects about gene regulation of skeletal muscle atrophy. The findings will not only enrich the molecular mechanism of denervation-induced muscle atrophy, but also provide potential targets for the prevention and treatment of denervation-induced muscle atrophy.
We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-21-1230).
Methods
Animal experiment
Animal experiments were carried out in accordance with the institutional animal care guidelines and approved by the Administration Committee of Experimental Animals, Jiangsu Province (20180305-004). Adult male Sprague-Dawley (SD) rats (weight, about 200 g) were provided by the Experimental Animal Center of Nantong University (Nantong, Jiangsu, China) with routine provision of food and water, at 22 °C, with a 12:12-h light-dark cycle. For animal treatments, SD rats were randomly divided into 2 groups (n=3), namely 1 sham group (only the sciatic nerve was exposed) and 1 experimental group (creating a 10 mm long defect of the sciatic nerve). After operation, all rats were euthanized under anesthesia at 14 d, and the TA muscles were dissected, rapidly frozen on liquid nitrogen, and stored at –80 °C for subsequent experiments.
Transcriptome sequencing analysis
Total RNA was extracted from muscle samples and its quality was detected. Then, the Truseq Stranded Total RNA with Ribo-Zero Gold kit (Illumina, San Diego, CA, USA) was used to digest the RNA into short pieces. The broken RNA was used as a template to synthesize first cDNA strand with random primers of 6 bases. Next, a second strand synthesis reaction system was formulated to synthesize second strand cDNA. During the synthesis of second strand cDNA, deoxyuridine triphosphate (dUTP) was substituted for deoxythymidine triphosphate (dTTP), and then different joints were connected. A strand containing dUTP was digested by uracil N-glycosylase (UNG) enzymatic method, and only 1 strand of cDNA connecting different joints of the strand was retained. A first strand of cDNA was purified using a purification kit. The end of the purified cDNA strand was repaired, a tail was added and the sequencing connector was connected, fragment size selection was performed, and polymerase chain reaction (PCR) amplification was performed at last. After the constructed RNA library had been qualified by Agilent 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA, USA), an Illumina sequencer was used for sequencing.
Screening of DEGs
The software DESeq (19) was used to standardize the counts of messenger RNA (mRNA) in each sample (Basemean value was used to estimate the expression level), and the difference multiple was calculated. A negative binomial (NB) distribution test was used to test the difference significance of the number of reads. Finally, the DEGs were screened according to the difference multiple and difference significance test results. The default screening difference condition was P<0.05 and the difference multiple was greater than 2.
Cluster analysis of DEGs
Unsupervised hierarchical clustering of DEGs was performed. We calculated the distance between pairs of multiple samples to form a distance matrix, combined the 2 closest classes into a new class, calculated the distance between the new class and the current class, and then combined and calculated until there was only 1 class. The expression of selected DEGs was used to calculate the direct correlation of samples. In general, the same sample could appear in the same cluster through clustering, and the genes clustered in the same cluster may have had similar biological functions.
GO enrichment analysis of DEGs
After the DEGs had been obtained, GO enrichment analysis was performed on the DEGs to describe their functions through the Database for Annotation, Visualization, and Integrated Discovery (DAVID) v6.8 (https://david.ncifcrf.gov/tools.jsp). The number of DEGs included in each GO item was counted, and the significance of DEG enrichment in each GO item was calculated by hypergeometric distribution test. The calculated results showed a P value of enrichment significance, with a small P value indicating enrichment of DEGs in the GO item. Those GO categories with a P value <0.05 were considered significantly enriched.
KEGG enrichment analysis of DEGs
The KEGG database (20) and DAVID bioinformatics resources were used for pathway analysis of DEGs, and hypergeometric distribution test was used to calculate the significance of DEGs enrichment in each pathway entry. The calculated results showed a P value of enrichment significance, with a small P value indicating enrichment of DEGs in the pathway entry. The KEGG pathway pictures were downloaded and the DEGs were labeled on the pathway map. Red indicated up-regulated genes, green indicated down-regulated genes, and yellow indicated both up-regulated and down-regulated genes. Those pathways with a P value <0.05 were considered significantly enriched.
Statistical analysis
When using RNA-seq data to compare and analyze whether there is differential expression of the same gene in two samples, two criteria can be selected: one is Fold Change, which is the change multiple of the same gene expression level in two samples; the other is P value or FDR (adjusted P value). In order to calculate the FDR value, we first calculate the P value of each gene, and then use the FDR error control method to correct the P value. The default screening condition was P<0.05 and the difference multiple was more than 2.
Results
DEGs in denervated skeletal muscles
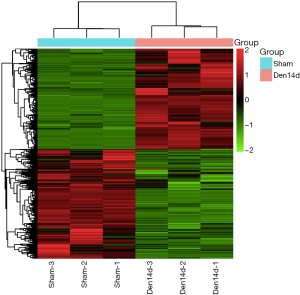
Transcriptome sequencing analysis was performed on the RNA sample extracted from TA muscles of rats at 0 and 14 days following sciatic nerve transection. Our study found that the expression of 5,690 transcripts in TA muscle changed at 14 days after denervation. Concretely, 2,749 transcripts were up-regulated, and 2,941 transcripts were down-regulated in denervated TA muscles after 14 days of denervation (Figure 1). These results in turn suggested that many biological processes and signaling pathways in skeletal muscle cells may also change significantly after denervation. These changes in biological processes and signaling pathways are bound to promote skeletal muscle atrophy.
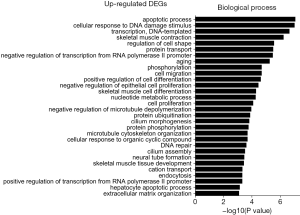
Biological process analysis
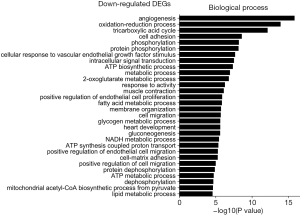
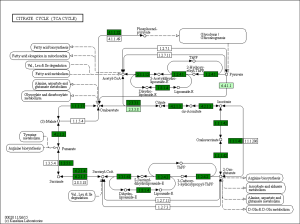
The GO was used to analyze biological processes related to DEGs, and the enriched categories of biological processes for the up-regulated expressed genes in TA muscles after 14 days of denervation are shown in Figure 2. Our results show that the up-regulated expressed genes mainly involve biological processes such as apoptotic process, cellular response to DNA damage stimulus, skeletal muscle contraction, aging, negative regulation of microtubule depolymerization, protein ubiquitination, and skeletal muscle tissue development. These data suggested that the apoptosis and ubiquitination proteolysis system was widely activated in denervated TA muscles after 14 days of denervation. The enriched categories of biological processes for the down-regulated genes in TA muscles after 14 days of denervation are labeled in Figure 3. As shown in Figure 3, the down-regulated expressed genes mainly involve in biological processes such as angiogenesis, oxidation-reduction process, tricarboxylic acid (TCA) cycle, cellular response to vascular endothelial growth factor stimulus, ATP biosynthetic process, muscle contraction, gluconeogenesis, nicotinamide adenine dinucleotide plus hydrogen (NADH) metabolic process, ATP synthesis coupled proton transport, positive regulation of endothelial cell migration, ATP metabolic process, and mitochondrial acetyl-CoA biosynthetic process from pyruvate. These results indicated that energy metabolism and angiogenesis are significantly inhibited in denervated skeletal muscle.
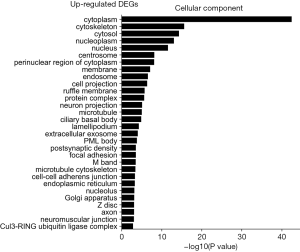
Cellular component analysis
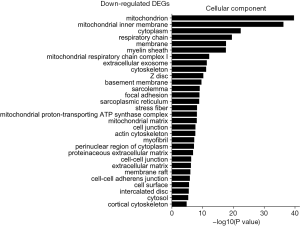
The GO was used to analyze the cellular component related to DEGs, and the enriched categories of cellular component for the up-regulated expressed genes in TA muscles after 14 days of denervation are labeled in Figure 4. As shown in Figure 4, the up-regulated expressed genes mainly involve in cellular components such as cytoplasm, cytoskeleton, cytosol, nucleoplasm, nucleus, and centrosome. These data suggested that the up-regulated expressed genes in denervated skeletal muscle mainly occurred in cytoplasm, cytoskeleton, and nucleus. The enriched categories of cellular component for the down-regulated genes in TA muscles after 14 days of denervation are shown in Figure 5. The down-regulated genes mainly occurred in cellular components such as mitochondrion, mitochondrial inner membrane, cytoplasm, respiratory chain, mitochondrial respiratory chain complex I, cytoskeleton, Z disc, sarcolemma, focal adhesion, mitochondrial proton-transporting ATP synthase complex, mitochondrial matrix, actin cytoskeleton, and myofibril. These data indicated that the down-regulated expressed genes mainly occurred in mitochondrion, cytoskeleton, and myofibril.
Molecular function analysis
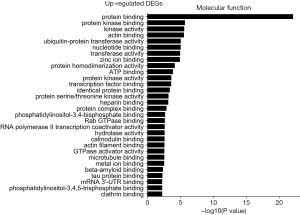
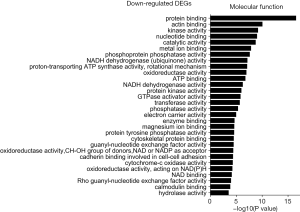
The GO was used to analyze the molecular function analysis related to DEGs, and the enriched categories of molecular function for the up-regulated expressed genes in TA muscles after 14 days of denervation are shown in Figure 6. Results showed that protein binding, actin binding, ubiquitin-protein transferase activity, transferase activity, ATP binding, transcription factor binding, protein serine/threonine kinase activity, hydrolase activity, calmodulin binding, and actin filament binding were enriched. These data suggested that the up-regulated expressed genes in denervated skeletal muscle mainly involve in protein binding, ubiquitin-protein transferase activity, and hydrolase activity. The enriched categories of molecular functions for the down-regulated genes in TA muscles after 14 days of denervation are displayed in Figure 7. As shown in Figure 7, the down-regulated expressed genes mainly involve the following molecular functions, such as protein binding, actin binding, catalytic activity, phosphoprotein phosphatase activity, NADH dehydrogenase (ubiquinone) activity, proton-transporting ATP synthase activity, oxidoreductase activity, ATP binding, NADH dehydrogenase activity, electron carrier activity, protein tyrosine phosphatase activity, cytoskeletal protein binding, cytochrome-c oxidase activity, and oxidoreductase activity. These data indicated that energy metabolism was significantly inhibited in TA muscles after denervation.
KEGG pathway enrichment analysis
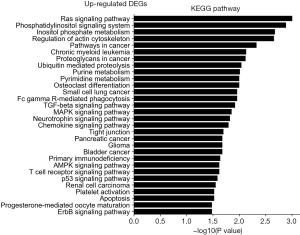
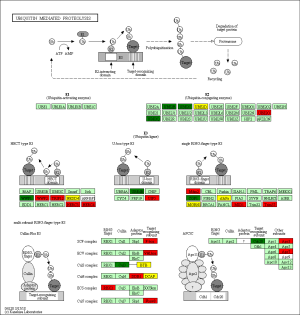
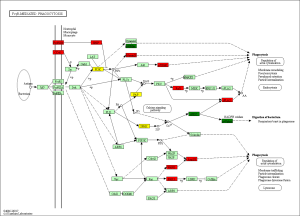
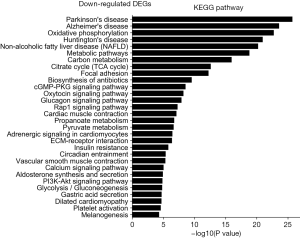
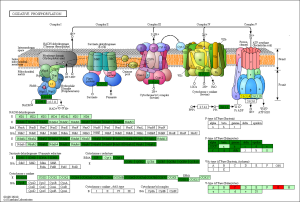
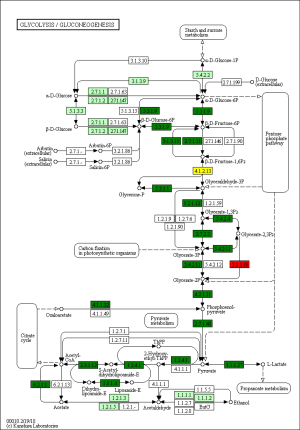
The KEGG pathway related to DEGs were identified through the DAVID database. The enriched categories of KEGG pathways for the up-regulated genes in TA muscles after 14 days of denervation are shown in Figure 8. Results showed that the Ras signaling pathway, ubiquitin mediated proteolysis, Fc gamma R-mediated phagocytosis, transforming growth factor-beta (TGF-β) signaling pathway, mitogen-activated protein kinase (MAPK) signaling pathway, chemokine signaling pathway, 5' AMP-activated protein kinase (AMPK) signaling pathway, T cell receptor signaling pathway, apoptosis, and p53 signaling pathway were enriched in denervated muscles. In particular, ubiquitin mediated proteolysis and Fc gamma R-mediated phagocytosis were significantly activated in in TA muscles after 14 days of denervation. As shown in Figure 9, the proteins involved in ubiquitin mediated proteolysis pathway, especially E3 ubiquitin ligases, were significantly up-regulated in denervated skeletal muscle. As shown in Figure 10, the proteins involved in Fc gamma R-mediated phagocytosis pathway were significantly up-regulated in TA muscles after 14 days of denervation. These results demonstrated that proteolysis pathway was significantly activated in denervated skeletal muscles. The activated TGF-β signaling pathway suggested that fibrosis plays an important role in the process of denervation-induced muscle atrophy. The enriched categories of KEGG pathways for the down-regulated genes in TA muscles after 14 days of denervation are labeled in Figure 11. As shown in Figure 12, most of the genes involved in oxidative phosphorylation were down-regulated in denervated skeletal muscle. As detailed in Figure 13, almost all the genes involved in TCA cycle were down-regulated in denervated skeletal muscle. Many genes involved in glycolysis/gluconeogenesis were down-regulated in denervated skeletal muscle (Figure 14). These results demonstrated that energy metabolism was significantly inhibited in TA muscles after 14 days of denervation. At the same time, the KEGG pathway involved in down-regulated genes included focal adhesion, the cGMP-PKG signaling pathway, Rap1 signaling pathway, cardiac muscle contraction, extracellular matrix (ECM)-receptor interaction, vascular smooth muscle contraction, calcium signaling pathway, and the PI3K-Akt signaling pathway in denervated muscles. These data suggested that protein synthesis was inhibited, and the function of muscle contract was decreased.
Discussion
The molecular mechanism of denervation atrophy remains unclear. The transcriptome sequencing analysis suggested that the expression of 5,690 transcripts in TA muscle changed at 14 days after denervation. A total of 2,749 transcripts were up-regulated, and 2,941 transcripts were down-regulated in denervated TA muscles after 14 days of denervation. The GO and KEGG enrichment analyses were used to analyze the DEGs, revealing some interesting aspects about gene regulation of skeletal muscle atrophy.
Biological process analysis showed that apoptosis and ubiquitination proteolysis system was widely activated in denervated TA muscles after 14 days of denervation. Cellular component analysis showed the up-regulated expressed genes in denervated skeletal muscle mainly occurred in cytoplasm, cytoskeleton, and nucleus. Molecular function analysis suggested that the up-regulated expressed genes in denervated skeletal muscle mainly involve in protein binding, ubiquitin-protein transferase activity, and hydrolase activity. The KEGG enrichment analysis showed that ubiquitin mediated proteolysis, Fc gamma R-mediated phagocytosis, TGF-β signaling pathway, MAPK signaling pathway, chemokine signaling pathway, AMPK signaling pathway, apoptosis, and p53 signaling pathway were enriched in denervated muscles. From GO and KEGG enrichment analyses, we found that the up regulated expressed genes mainly occur in cytoplasm and cytoskeleton, have ubiquitin protein transferase activity and hydrolase activity, participate in ubiquitination proteolysis, and involve mainly ubiquitin mediated proteolysis, Fc gamma R-mediated phagocytosis, and TGF-ß signaling pathway. These results are consistent with those of previous studies. The UPS is important for muscle homeostasis and health (21). Now, many studies have shown that UPS impairment affects not only matured muscle fibers, but also muscle stem cells (21). This ubiquitination enzymatic cascade, including ubiquitin E1 (activating enzyme), ubiquitin E2 (conjugating enzyme), and ubiquitin E3 (ligase), is very important for the activation of UPS. The UPS is one of the major pathways that mediate proteolysis in muscle. Many UPS genes were activated in skeletal muscle atrophy (22). Atrogin-1 and MuRF-1 are 2 muscle specific E3 ubiquitin ligases that are important regulators of ubiquitin-proteasome system in skeletal muscle (23,24). Low nutrients, disuse, systemic inflammation, unloading, chronic kidney disease (CKD), cancer, or ageing provoke a catabolic state characterized by enhanced muscle proteolysis (25,26). Our previous study also found that the expression of Atrogin-1 and MuRF-1 was significantly increased in denervated skeletal muscle, which played an important role in the degradation of muscle protein (11,14,18). The sequencing results of this study found that ubiquitin-conjugating enzyme E2O (UBE2O), WW domain containing E3 ubiquitin protein ligase 2 (WWP2), thyroid hormone receptor interactor 12 (TRIP12), HECT and RLD domain containing E3 ubiquitin protein ligase 3/4 (HERC3/4), ubiquitin-conjugating enzyme 7- interacting protein 5 (UIP5), MDM2 proto-oncogene (Mdm2), midline 1 (MID1), and tripartite motif-containing 37 (Trim37) were up-regulated in denervated skeletal muscles, which confirmed the important role of ubiquitin-mediated proteolysis in the process of denervation-induced muscle atrophy. Berardi et al. found that increased auto-phagocytosis results in muscle wasting in cachexia (27). Recently, lysosomes, containing many hydrolases and proteases, have been considered to play an important role in maintaining tissue homeostasis, mainly through autophagy, phagocytosis, and endocytosis (28). The ALS plays a key role in protein degradation in skeletal muscle cells (29,30). It is widely activated during muscle atrophy and leads to muscle mass loss in varying degrees (28). Our previous study also found that ALS is widely activated in the process of denervation-induced muscle atrophy (6,12,14). The results of this study also confirm our previous results. Our study showed that ubiquitin-proteasome system and autophagy lysosomal system are activated in the process of denervation-induced muscle atrophy, but the trigger factors that initiate their activation are not yet clear. Our previous studies suggested that oxidative stress and inflammation may be the trigger factors for activation of the ubiquitin-proteasome and autophagy lysosomal systems (6,18,31). Further studies are needed to confirm the possible molecular mechanism.
Biological process analysis showed that angiogenesis, oxidation-reduction process, TCA cycle, ATP biosynthetic process, muscle contraction, gluconeogenesis, and NADH metabolic process was inhibited in denervated TA muscles after 14 days of denervation. Cellular component analysis showed the down-regulated expressed genes in denervated skeletal muscle mainly occurred in mitochondrion, cytoskeleton, and myofibril. Molecular function analysis suggested that the down-regulated expressed genes in denervated skeletal muscle mainly involve in actin binding, catalytic activity, NADH dehydrogenase (ubiquinone) activity, proton-transporting ATP synthase activity, oxidoreductase activity, ATP binding, NADH dehydrogenase activity, and electron carrier activity. These data indicated that energy metabolism was significantly inhibited in TA muscles after denervation. The enriched categories of KEGG pathways for the down-regulated genes in TA muscles after 14 days of denervation included oxidative phosphorylation, TCA cycle, glycolysis/gluconeogenesis, PI3K-Akt signaling pathway, and focal adhesion. From GO and KEGG enrichment analyses, we found that the down-regulated expressed genes mainly occur in mitochondrion, cytoskeleton, and myofibril, have catalytic activity, NADH dehydrogenase (ubiquinone) activity, proton-transporting ATP synthase activity, oxidoreductase activity, participate in energy metabolism and protein synthesis, and mainly involve oxidative phosphorylation, TCA cycle, and glycolysis/gluconeogenesis. These results indicated that energy metabolism was significantly inhibited in TA muscles after 14 days of denervation. Mitochondria are most commonly known for energy production by oxidative phosphorylation (32), and are the primary provider of ATP, which is essential for physiological muscle activities (33). Mitochondria have been thought to contribute to skeletal myopathy (34). Dysfunctional mitochondria can impair cells by decreased ATP production and increased oxidative stress (35). Migliavacca et al. found that individuals with sarcopenia demonstrated a prominent transcriptional signature of mitochondrial bioenergetic dysfunction in skeletal muscle, reduced mitochondrial respiratory complex expression, and downregulation of oxidative phosphorylation in sarcopenic muscle (36). Hughes et al. suggested that mitochondrial bioenergetic dysfunctions were associated with induction of markers of muscle atrophy in some muscles, including Duchenne muscular dystrophy (DMD) (37). Mitochondrial metabolism was disrupted in skeletal muscles from patients with CKD (38). A mouse model of CKD displayed significant reductions in mitochondrial oxidative phosphorylation, which was associated with progressive skeletal atrophy, weakness, and fatigue (34). Thome et al. suggested that disruption of mitochondrial oxidative phosphorylation was confirmed by decreased respiratory capacity and elevated superoxide production in cultured myotubes (39). Mitochondrial dysfunction may play a significant role in the etiology of Huntington’s disease, which led to severe clinical impairments-skeletal muscle atrophy (40). Gouspillou et al. demonstrated that mitochondrial dysfunction was implicated in skeletal muscle atrophy (41). Mitochondrial energy metabolism [TCA cycle and oxidative phosphorylation] was significantly inhibited in skeletal muscles from sarcopenia (42). In conclusion, the function of the involved down-regulated genes involved mainly included energy metabolism during denervation-induced muscle atrophy. This study further confirmed that abnormal mitochondrial energy metabolism may play an important role in the process of muscle atrophy. As for the specific causes of abnormal mitochondrial energy metabolism in atrophied muscles, further research is needed.
In conclusion, there are a multitude of DEGs in skeletal muscle after denervation. The DEGs mainly involve in proteolysis, apoptosis, ageing, energy metabolism, angiogenesis, and protein synthesis. This study not only enriched the molecular regulation mechanism of denervation-induced muscle atrophy, but also provided a potential molecular target for the prevention and treatment of muscle atrophy.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (Nos. 82072160 and 81901933), the Major Natural Science Research Projects in Universities of Jiangsu Province (No. 20KJA310012), Nantong Social Development Fund (MSZ19179), and Nantong University Doctoral Research Start up Fund project (18B15).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-1230
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-1230
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-1230). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Animal experiments were carried out in accordance with the institutional animal care guidelines and approved by the Administration Committee of Experimental Animals, Jiangsu Province (20180305-004).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Midrio M. The denervated muscle: facts and hypotheses. A historical review. Eur J Appl Physiol 2006;98:1-21. [Crossref] [PubMed]
- Yang X, Xue P, Liu X, et al. HMGB1/autophagy pathway mediates the atrophic effect of TGF-beta1 in denervated skeletal muscle. Cell Commun Signal 2018;16:97. [Crossref] [PubMed]
- Reza MM, Subramaniyam N, Sim CM, et al. Irisin is a pro-myogenic factor that induces skeletal muscle hypertrophy and rescues denervation-induced atrophy. Nat Commun 2017;8:1104. [Crossref] [PubMed]
- Gu X, Ding F, Yang Y, et al. Construction of tissue engineered nerve grafts and their application in peripheral nerve regeneration. Prog Neurobiol 2011;93:204-30. [Crossref] [PubMed]
- Shen Y, Zhang R, Xu L, et al. Microarray Analysis of Gene Expression Provides New Insights Into Denervation-Induced Skeletal Muscle Atrophy. Front Physiol 2019;10:1298. [Crossref] [PubMed]
- Shen Y, Zhang Q, Huang Z, et al. Isoquercitrin Delays Denervated Soleus Muscle Atrophy by Inhibiting Oxidative Stress and Inflammation. Front Physiol 2020;11:988. [Crossref] [PubMed]
- Fedorov VB, Goropashnaya AV, Stewart NC, et al. Comparative functional genomics of adaptation to muscular disuse in hibernating mammals. Mol Ecol 2014;23:5524-37. [Crossref] [PubMed]
- Jeong HO, Park D, Im E, et al. Determination of the Mechanisms that Cause Sarcopenia through cDNA Microarray. J Frailty Aging 2017;6:97-102. [PubMed]
- Krüger K, Dischereit G, Seimetz M, et al. Time course of cigarette smoke-induced changes of systemic inflammation and muscle structure. Am J Physiol Lung Cell Mol Physiol 2015;309:L119-28. [Crossref] [PubMed]
- Weng J, Zhang P, Yin X, et al. The Whole Transcriptome Involved in Denervated Muscle Atrophy Following Peripheral Nerve Injury. Front Mol Neurosci 2018;11:69. [Crossref] [PubMed]
- Huang Z, Zhong L, Zhu J, et al. Inhibition of IL-6/JAK/STAT3 pathway rescues denervation-induced skeletal muscle atrophy. Ann Transl Med 2020;8:1681. [Crossref] [PubMed]
- Huang Z, Fang Q, Ma W, et al. Skeletal Muscle Atrophy Was Alleviated by Salidroside Through Suppressing Oxidative Stress and Inflammation During Denervation. Front Pharmacol 2019;10:997. [Crossref] [PubMed]
- Ma W, Zhang R, Huang Z, et al. PQQ ameliorates skeletal muscle atrophy, mitophagy and fiber type transition induced by denervation via inhibition of the inflammatory signaling pathways. Ann Transl Med 2019;7:440. [Crossref] [PubMed]
- Qiu J, Fang Q, Xu T, et al. Mechanistic Role of Reactive Oxygen Species and Therapeutic Potential of Antioxidants in Denervation- or Fasting-Induced Skeletal Muscle Atrophy. Front Physiol 2018;9:215. [Crossref] [PubMed]
- Qiu J, Wu L, Chang Y, et al. Alternative splicing transitions associate with emerging atrophy phenotype during denervation-induced skeletal muscle atrophy. J Cell Physiol 2021;236:4496-514. [Crossref] [PubMed]
- Qiu J, Zhu J, Zhang R, et al. miR-125b-5p targeting TRAF6 relieves skeletal muscle atrophy induced by fasting or denervation. Ann Transl Med 2019;7:456. [Crossref] [PubMed]
- Sun H, Gong Y, Qiu J, et al. TRAF6 inhibition rescues dexamethasone-induced muscle atrophy. Int J Mol Sci 2014;15:11126-41. [Crossref] [PubMed]
- Wan Q, Zhang L, Huang Z, et al. Aspirin alleviates denervation-induced muscle atrophy via regulating the Sirt1/PGC-1alpha axis and STAT3 signaling. Ann Transl Med 2020;8:1524. [Crossref] [PubMed]
- Anders S, Huber W. Differential Expression of RNA-Seq Data at the Gene Level-The DESeq Package. European Molecular Biology Laboratory (EMBL), Heidelberg, 10: f1000research 2012.
- Pertea M, Pertea GM, Antonescu CM, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 2015;33:290-5. [Crossref] [PubMed]
- Kitajima Y, Yoshioka K, Suzuki N. The ubiquitin-proteasome system in regulation of the skeletal muscle homeostasis and atrophy: from basic science to disorders. J Physiol Sci 2020;70:40. [Crossref] [PubMed]
- Khalil R. Ubiquitin-Proteasome Pathway and Muscle Atrophy. Adv Exp Med Biol 2018;1088:235-48. [Crossref] [PubMed]
- Gumucio JP, Mendias CL. Atrogin-1, MuRF-1, and sarcopenia. Endocrine 2013;43:12-21. [Crossref] [PubMed]
- Huang Z, Zhu J, Ma W, et al. Strategies and potential therapeutic agents to counter skeletal muscle atrophy. Biotarget 2018;2:8. [Crossref]
- Milan G, Romanello V, Pescatore F, et al. Regulation of autophagy and the ubiquitin-proteasome system by the FoxO transcriptional network during muscle atrophy. Nat Commun 2015;6:6670. [Crossref] [PubMed]
- Wang XH, Mitch WE. Mechanisms of muscle wasting in chronic kidney disease. Nat Rev Nephrol 2014;10:504-16. [Crossref] [PubMed]
- Berardi E, Aulino P, Murfuni I, et al. Skeletal muscle is enriched in hematopoietic stem cells and not inflammatory cells in cachectic mice. Neurol Res 2008;30:160-9. [Crossref] [PubMed]
- Gumpper K, Sermersheim M, Zhu MX, et al. Skeletal Muscle Lysosomal Function via Cathepsin Activity Measurement. Methods Mol Biol 2019;1854:35-43. [Crossref] [PubMed]
- Franekova V, Storjord HI, Leivseth G, et al. Protein homeostasis in LGMDR9 (LGMD2I) - The role of ubiquitin-proteasome and autophagy-lysosomal system. Neuropathol Appl Neurobiol 2020; Epub ahead of print. [Crossref] [PubMed]
- Manickam R, Duszka K, Wahli W. PPARs and Microbiota in Skeletal Muscle Health and Wasting. Int J Mol Sci 2020;21:8056. [Crossref] [PubMed]
- Ma W, Xu T, Wang Y, et al. The role of inflammatory factors in skeletal muscle injury. Biotarget 2018;2:7. [Crossref]
- Christian BE. Mitochondrial ribosome-binding factor A (mtRBFA): mediator of rRNA methylation and maturation. Biotarget 2017;1:16. [Crossref]
- Li A, Yi J, Li X, et al. Physiological Ca(2+) Transients Versus Pathological Steady-State Ca(2+) Elevation, Who Flips the ROS Coin in Skeletal Muscle Mitochondria. Front Physiol 2020;11:595800 [Crossref] [PubMed]
- Thome T, Kumar RA, Burke SK, et al. Impaired muscle mitochondrial energetics is associated with uremic metabolite accumulation in chronic kidney disease. JCI Insight 2020;6:e139826 [Crossref] [PubMed]
- Thelen MP, Wirth B, Kye MJ. Mitochondrial defects in the respiratory complex I contribute to impaired translational initiation via ROS and energy homeostasis in SMA motor neurons. Acta Neuropathol Commun 2020;8:223. [Crossref] [PubMed]
- Migliavacca E, Tay SKH, Patel HP, et al. Mitochondrial oxidative capacity and NAD(+) biosynthesis are reduced in human sarcopenia across ethnicities. Nat Commun 2019;10:5808. [Crossref] [PubMed]
- Hughes MC, Ramos SV, Turnbull PC, et al. Early myopathy in Duchenne muscular dystrophy is associated with elevated mitochondrial H2 O2 emission during impaired oxidative phosphorylation. J Cachexia Sarcopenia Muscle 2019;10:643-61. [Crossref] [PubMed]
- Rao M, Jaber BL, Balakrishnan VS. Chronic kidney disease and acquired mitochondrial myopathy. Curr Opin Nephrol Hypertens 2018;27:113-20. [Crossref] [PubMed]
- Thome T, Salyers ZR, Kumar RA, et al. Uremic metabolites impair skeletal muscle mitochondrial energetics through disruption of the electron transport system and matrix dehydrogenase activity. Am J Physiol Cell Physiol 2019;317:C701-13. [Crossref] [PubMed]
- Rodinova M, Krizova J, Stufkova H, et al. Deterioration of mitochondrial bioenergetics and ultrastructure impairment in skeletal muscle of a transgenic minipig model in the early stages of Huntington's disease. Dis Model Mech 2019;12:dmm038737 [Crossref] [PubMed]
- Gouspillou G, Sgarioto N, Kapchinsky S, et al. Increased sensitivity to mitochondrial permeability transition and myonuclear translocation of endonuclease G in atrophied muscle of physically active older humans. FASEB J 2014;28:1621-33. [Crossref] [PubMed]
- Ibebunjo C, Chick JM, Kendall T, et al. Genomic and proteomic profiling reveals reduced mitochondrial function and disruption of the neuromuscular junction driving rat sarcopenia. Mol Cell Biol 2013;33:194-212. [Crossref] [PubMed]
(English Language Editor: J. Jones)


