
G-rich sequence factor 1 serves as a prognostic biomarker in septic patients
Introduction
Sepsis is a systemic disease, which deteriorates rapidly with multiple organ dysfunction (1). In the United States, despite advanced medical resources, the mortality rate of sepsis is 34–56%, according to recent retrospective clinical studies (2). Sepsis is not only a major health issue but also a serious economic problem (3). It is considered to be a systemic response of infection including bacteria, viruses, and fungi (4,5). Inflammatory factors such as tumor necrosis factor α (TNF-α), interleukin 6 (IL-6) and anti-inflammatory factors such as interleukin 10 (IL-10) (6,7) are involved in the development of sepsis. Macrophage dysfunction has been shown to be associated with exacerbation of sepsis (8-10).
Some RNA-binding proteins (RBPs) are related to multiple functions of eukaryotic cells (11), including mobilization, translation, and apoptosis., As an RBP with G-rich elements, G-rich sequence factor 1 (GRSF1) (12) participates in essential cellular processes in senescent cells (13,14). It is also considered a mitochondrial protein (14) involved in stabilizing target messenger RNAs (mRNAs). Lipopolysaccharide (LPS) induces inflammatory responses by activating nuclear factor kappa light chain enhancer of activated B cells (NF-κB) signaling pathways, and is the main element of gram-negative bacteria cell walls (7,15,16). Loss of GRSF1 is associated with increased production of the pro-inflammatory factor IL-6 (17). Over production and secretion of pro-inflammatory factors is a vital sign of sepsis deterioration (18). These findings have indicated that GRSF1 may play an important role in the inflammatory response of sepsis. We hypothesized that GRSF1 could be a potential biomarker in the prognosis of septic patients.
In our study, expression of GRSF1 in the peripheral blood of septic patients and healthy controls were compared, and the severity of sepsis was evaluated by combined GRSF1 and other biomarkers. We used RAW 264.7 cells and mouse peritoneal macrophages induced by LPS to investigated GRSF1 expression in vitro, and cecal ligation and puncture (CLP) induced mice were established to explore GRSF1 expression in vivo. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-21-1022).
Methods
Participant cohort and healthy donors
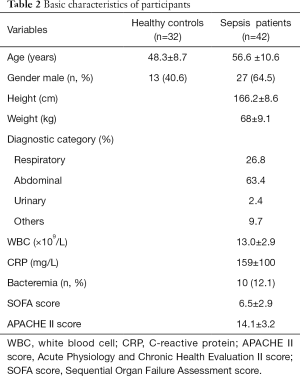
A total of 42 sepsis patients according to the definitions of the Sepsis 3.0 2016 (19) between 1 April 2019 and 31 December 2019 in our hospital and 32 healthy donors were included in this study. Blood samples were collected within 24 h of emergency department admission. The Sequential Organ Failure Assessment (SOFA) and Acute Physiologic Assessment and Chronic Health Evaluation II (APACHE II) scores were calculated, white blood cells (WBC), and C-reactive protein (CRP) were recorded at the time of hospitalization. This study protocol was approved by the Ethics Committee of the Affiliated Hospital of Nantong University (2017-L021) with conformation to the Declaration of Helsinki (as revised in 2013). Participants or authorized clients provided informed consent prior to commencement of the study.
CLP induced septic mice
Female C57BL/6J mice, aged 6–8 weeks, and weighing about 22 g, were obtained from the Laboratory Animal Center of Nantong University. A mouse model of cecal ligation and perforation (CLP) induced sepsis was established. Mice were randomly subdivided into a sham operation group and sepsis group (n=6). Before surgical procedures, mice were anesthetized with 10% chloral hydrate (400 mg/kg) via intraperitoneal injection. A mid-line laparotomy incision (1–2 cm) was made to expose the cecum. Then, 50% of the cecum was ligated and a single hole was perforated with an 18-gauge needle. Next, the cecum was replaced in the abdominal cavity, and the incision was closed. The sham group underwent a similar procedure without ligation or perforation. Mice were checked every 8 hours for survival analysis. All animal experiments were undertaken in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals. Animal experiments were approved by Ethics Committee of Affiliated Hospital of Nantong University.
Isolation and purification of mouse peritoneal macrophages
The mice were purchased from the Laboratory Animal Center of Nantong University and housed according to standard protocol. To isolate and purify macrophages from mouse peritoneum, 1 mL of 6% starch broth was injected into the peritoneum before collection. After 2–3 days, the mice were euthanized and their abdomens were disinfected with 75% alcohol. The outer layer of the mice peritonea were cut open with scissors and 10 mL cold phosphate buffered saline (PBS) was injected into the abdominal cavity in order to wash the peritoneal macrophages. The peritoneum was gently massaged after the injection of PBS to exfoliate any attached cells into the fluid. The PBS was collected into a 15 mL tube from the peritoneum with a sterile straw and centrifuged at 1,000 ×g for 5 min. After centrifugation, the supernatant was removed and the cells were cultured in RPMI-1640 combined with 100 U/mL penicillin, 100 mg/mL streptomycin, and 10% fetal bovine serum [(FBS); cat. No. E600001; Sangon Biotech, Shanghai, China], at 37 °C and 5% CO2. The cells were seeded into a 6-well plate (1×106 cells/well) and incubated overnight. Non-adherent cells were gently washed 3 times with warm PBS. After completing the above experimental steps, about 90% of the pure macrophages were used in the experiment.
Stimulation with LPS
Mouse peritoneal macrophages were seeded into a 6-well plate (1×106 cells/well), and stimulated for 2, 4, 8, 16, and 24 hours with 100 ng/mL Escherichia coli LPS (InvivoGen, San Diego, CA, USA). Cells without LPS stimulation served as a control group. About 5×105 cells per well were seeded in a 6-well plate and then stimulated with 100 ng/mL LPS for 2, 4, 8, 16, and 24 hours. Cells without treatment with LPS served as a control group.
Reverse transcription polymerase chain reaction (RT-PCR)
Total RNA was extracted from macrophages or tissues using TRIzol™ Reagent (Invitrogen, Carlsbad, CA, USA). HiScript® II Q Select RT SuperMix for qPCR (+gDNA wiper) (Vazyme, Nanjing, China) was purchased for RT-PCR, and performed according to the manufacturers. The resulting complementary DNA (cDNA) was used for qPCR detection using the StepOnePlus qPCR system (Applied Biosystems, Waltham, MA, USA) with ChamQ Universal SYBR qPCR Master Mix (Vazyme). Quantitative evaluation of target gene expression was performed using the ΔΔCT method. The expression of GRSF1 was normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) expression. Primer sequences are provided in Table 1.
Full table
Western blotting
Tissues (50 mg) were prepared with radio immunoprecipitation assay (RIPA) combined with protease inhibitors (ab65621, Abcam, Burlingame, USA) and 100 uM phenylmethylsulfonyl fluoride (PMSF) (#P7626, Sigma). After homogenization, the mixture was incubated on ice for 30 min. The supernatant was collected, followed by centrifugation at 4 °C for 10 minutes at 14,000 g. A DC Bradford assay kit (Bio-Rad, Hercules, CA, USA) was used to measure the protein concentration. Protein (200 ng) was electrophoresed in an 8–12% Tris-Glycine gel (Sangon Biotech) and electrotransferred onto a polyvinylidene difluoride (PVDF) membrane (Thermo Fisher Scientific). In order to eliminate the influence of nonspecific protein binding, the membrane was blocked with 5% nonfat milk, dissolved in 0.05% tris-buffered saline and Tween 20 (TBST), at room temperature for at least 1 h. The membranes were incubated with primary antibodies against GRSF1 (1:1,000; Abcam) and β-actin (1:4,000; Abcam) overnight at 4 °C. After incubation with secondary antibodies (Thermo Fisher Scientific), western blotting signals were visualized with electrochemiluminescence (ECL) or ECL prime by Bio-Rad Gel Doc XR+ (Bio-Rad). The stripe density was normalized with ImageJ v1.47 software (National Institutes of Health, Bethesda, MD, USA).
Statistical analysis
The software GraphPad Prism 5.0 (GraphPad Software, Inc., San Diego, CA, USA) was used for data analysis. Statistical significance was tested by Student’s t-test in pairwise comparison. All data were presented as mean ± standard deviation (SD) or median with interquartile range (IQR) depending on the test. The level of statistical significance was set at P<0.05.
Results
GRSF1 expression in patients with sepsis
The expression of GRSF1 was analyzed among 42 sepsis patients admitted to the Emergency Department of the Affiliated Hospital of Nantong University. When compared, the expression of GRSF1 was shown to be incredibly lower in sepsis patients than HC (mean 0.4449±0.0516 vs. 1.204±0.1438, P<0.0001) (Figure 1).
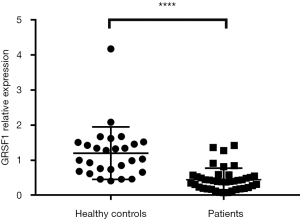
The basic characteristics of comparison between sepsis patients and healthy controls are shown in Table 1. The 42 participants were divided into 2 groups by quartile, with high expression in one group and lower in the other. The characteristics comparisons are shown in Table 2 and Table 3.
Full table
Full table
Correlation of GRSF1 expression with clinical characteristics of sepsis patients
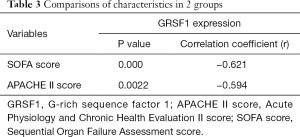
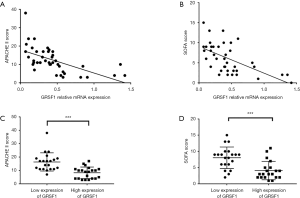
Considering all the clinical characteristics of patients in sepsis, surprisingly, GRSF1 expression was negatively related to the APACHE II score and SOFA scores (Table 4, Figure 2 A,B). Contrarily, height, weight, age, WBC, and CRP were not associated with GRSF1 expression. The APACHE II score of the group of low expression of GRSF1 was significantly higher than that of the group of high expression of GRSF1 (Figure 2C). The SOFA score of the group of low expression of GRSF1 was significantly higher than that of the group of high expression of GRSF1 (Figure 2D). Thus, GRSF1 expression may be negatively related to the severity of sepsis.
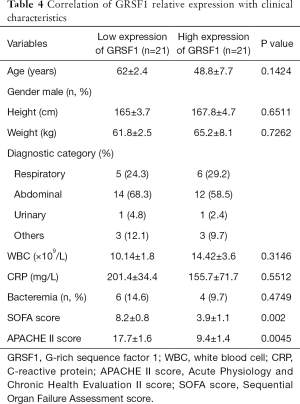
Full table
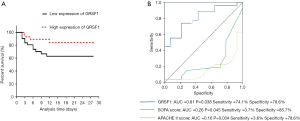
Prognostic value of the expression of GRSF1 in patients with sepsis
The 28-day survival rate of patients with the group of low expression of GRSF1 and the group of high expression of GRSF1 was analyzed, to find out the relationship between the expression of GRSF1 in the peripheral blood of patients with sepsis and the prognosis of sepsis. The 28-day survival rate of patients in the group of low expression of GRSF1 was significantly lower than that of patients with the group of high expression of GRSF1 (Figure 3A). It is shown that the survival rate of the group of low GRSF1 expression was 62.9% and that of the group of high GRSF1 expression was 84.1%. Combined with the expression of GRSF1 and the 28-day survival rate were analyzed by the ROC curve analysis. The results showed that GRSF1 had a certain predictive ability for the 28-day survival rate of patients with sepsis (AUC =0.688, P=0.040). Meanwhile, SOFA score and APACHE II score and 28-day survival rate were analyzed by the ROC curve (Figure 3B). According to the analysis of the 28-day survival rate, it is suggested that the expression of GRSF1 in peripheral blood of patients with sepsis plays a great value for early diagnosis.
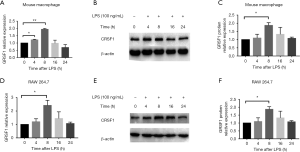
The GRSF1 expression in RAW 264.7 Cells and mouse peritoneal macrophages after LPS-stimulation
We aimed to explore the role of GRSF1 in the inflammatory response of macrophages stimulated by LPS. Firstly, in RAW 264.7 cells were stimulated by 100 ng/mL LPS for the indicated times, and we then examined the expression of GRSF1. At the mRNA level, GRSF1 significantly increased at 8 h and then gradually decreased to the normal level within 24 h (Figure 4A). In protein analysis, GRSF1 had increased at 8 and 16 h (Figure 4B,C). In addition, we also analyzed GRSF1 expression in mouse peritoneal macrophages under the same conditions. The GRSF1 mRNA level was increased at 4 and 8 h and then slightly decreased within 24 h (Figure 4D). The GRSF1 protein level was increased at 4 and 8 h (Figure 4E,F).
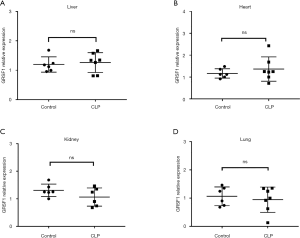
The expression of GRSF1 in sepsis in the CLP model in different organs
To evaluate the role of GRSF1 in sepsis, we performed a CLP surgery in mice to induce sepsis. The surgery was performed according to protocol (20). We evaluated this by performing qPCR for GRSF1 using RNA extracted from mouse heart, liver, lung, and kidney (Figure 5A,B,C,D), but the differences were not statistically significant.
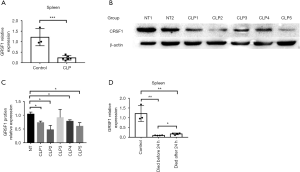
GRSF1 was decreased in sepsis in the CLP model, especially in the spleen
Compared to sham operated mice, decreased expression of GRSF1 was observed in the spleen of mice treated with CLP (Figure 6A,B,C). These data suggest that the expression of GRSF1 is decreased in the spleen in CLP-induced sepsis mice. To determine whether expression in the spleen was associated with the severity of sepsis in mice, we divided the CLP models into 2 groups according to whether the mice survived beyond 24 hours. The GRSF1 expression in mice that died before 24 hours was much lower than in mice that survived much longer than 24 hours (Figure 6D). Therefore, the spleen was shown to be the target organ of GRSF1. Under these circumstances, we could speculate that the lower the GRSF1 expression in sepsis, the more serious the severity of sepsis.
Discussion
Our previous study had already shown that GRSF1 expression in patients with sepsis differed from that of HC, according to the peripheral blood of patients with sepsis and HC. We found that GRSF1 expression was markedly lower in sepsis patients compared with healthy individuals.
Although GRSF1 expression in inflammation or sepsis in humans has not been studied extensively, it has been found that GRSF1 is a mitochondrial protein which regulates RNA processing. Until now, the physiological function of GRSF1 is still unclear; however, some studies have indicated possible pathologies related to inflammation. In CLP mice, we found that the protective effects depended on GRSF1 expression. We will explore the mechanism of GRSF1 in sepsis in future research.
In this study, we also discovered a negative correlation between the SOFA and APACHE II scores and GRSF1 expression. It is widely known that the SOFA and APACHE II scores are markers to predict the severity of sepsis (21,22). Simultaneously, our findings revealed that both the SOFA and APACHE II scores were negatively associated with GRSF1 expression. Given the contrary relationship of the SOFA and APACHE II scores and GRSF1 expression, our study suggested that GRSF1 expression in vivo contributes to protection from death during sepsis. In CLP model mice, it was demonstrated that GRSF1 may play an essential role in protecting mice from earlier death. Thus, our clinical data are consistent with the animal study. In conclusion, these findings suggested that GRSF1 expression on peripheral blood could be considered for evaluating the severity of sepsis.
Our research had several limitations. Firstly, considering that the number of cases included in this study was insufficient, subgroup analysis was to study the effects of admission, complications, or infection sites on GRSF1 expression was not feasible. Secondly, the research was performed in a single hospital center, in this case, the scalability can only be set to suit a limited variety of situations. Thirdly, the study lacked correlation with GRSF1 expression and 28-day mortality. Fourthly, in this study, we aimed to confirm whether GRSF1 expression could be used as a biomarker for early evaluation of severity in sepsis patients. Thus we measured the GRSF1 expression in peripheral blood within a 24 h period, instead of consistent measurement. For further research, we need to ascertain if changes in GRSF1 expression are related to the SOFA and APACHE II scores.
Conclusions
Considering the SOFA and APACHE II scores are predictors of sepsis severity, together with the correlation between GRSF 1 expression, GRSF1 are likely to be considered as a promising biomarker of the sepsis severity.
Acknowledgments
The authors thank The Affiliated Hospital of Nantong University, for expert technical assistance and statistical analysis support. The manuscript is approved by all authors for publication.
Funding: This research received funding from the National Natural Science Foundation of China Youth Found (81801893) and Nantong Clinical Medicine Research Center (HS2019005 and HS2020001), and the Nantong scientific projects (MS12020006 and MS12020017).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-1022
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-1022
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-1022). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethics Committee of the Affiliated Hospital of Nantong University (2017-L021) with conformation to the Declaration of Helsinki (as revised in 2013). Participants or authorized clients provided informed consent prior to commencement of the study. All animal experiments were undertaken in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals. Animal experiments were approved by Ethics Committee of Affiliated Hospital of Nantong University.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Fleischmann C, Scherag A, Adhikari NK, et al. Assessment of Global Incidence and Mortality of Hospital-treated Sepsis. Current Estimates and Limitations. Am J Respir Crit Care Med 2016;193:259-72. [Crossref] [PubMed]
- Wu CC, Lan HM, Han ST, et al. Comparison of diagnostic accuracy in sepsis between presepsin, procalcitonin, and C-reactive protein: a systematic review and meta-analysis. Ann Intensive Care 2017;7:91. [Crossref] [PubMed]
- Brodska H, Valenta J, Pelinkova K, et al. Diagnostic and prognostic value of presepsin vs. established biomarkers in critically ill patients with sepsis or systemic inflammatory response syndrome. Clin Chem Lab Med 2018;56:658-68. [Crossref] [PubMed]
- Sen P, Demirdal T, Nemli SA, et al. Diagnostic and prognostic value of new bioscore in critically ill septic patients. Arch Physiol Biochem 2019; Epub ahead of print. [Crossref] [PubMed]
- Park SB, Park GH, Um Y, et al. Wood-cultivated ginseng exerts anti-inflammatory effect in LPS-stimulated RAW264.7 cells. Int J Biol Macromol 2018;116:327-34. [Crossref] [PubMed]
- Arefian H, Heublein S, Scherag A, et al. Hospital-related cost of sepsis: A systematic review. J Infect 2017;74:107-17. [Crossref] [PubMed]
- Kondo Y, Umemura Y, Hayashida K, et al. Diagnostic value of procalcitonin and presepsin for sepsis in critically ill adult patients: a systematic review and meta-analysis. J Intensive Care 2019;7:22. [Crossref] [PubMed]
- Jhan JY, Huang YT, Shih CH, et al. Procalcitonin levels to predict bacterial infection in surgical intensive care unit patients. Formos J Surg 2017;50:135-41.
- Huang X, Venet F, Wang YL, et al. PD-1 expression by macrophages plays a pathologic role in altering microbial clearance and the innate inflammatory response to sepsis. Proc Natl Acad Sci U S A 2009;106:6303-8. [Crossref] [PubMed]
- Li J, Chen J, Kirsner R. Pathophysiology of acute wound healing. Clin Dermatol 2007;25:9-18. [Crossref] [PubMed]
- Zhang Z, Yang T, Xiao J. Circular RNAs: Promising Biomarkers for Human Diseases. EBioMedicine 2018;34:267-74. [Crossref] [PubMed]
- Nishida K, Kuwano Y, Nishikawa T, et al. RNA Binding Proteins and Genome Integrity. Int J Mol Sci 2017;18:1341. [Crossref] [PubMed]
- Jourdain AA, Koppen M, Wydro M, et al. GRSF1 regulates RNA processing in mitochondrial RNA granules. Cell Metab 2013;17:399-410. [Crossref] [PubMed]
- Antonicka H, Sasarman F, Nishimura T, et al. The Mitochondrial RNA-Binding Protein GRSF1 Localizes to RNA Granules and Is Required for Posttranscriptional Mitochondrial Gene Expression. Cell Metabolism 2013;17:386-98. [Crossref] [PubMed]
- Zou YH, Zhao L, Xu YK, et al. Anti-inflammatory sesquiterpenoids from the Traditional Chinese Medicine Salvia plebeia: Regulates pro-inflammatory mediators through inhibition of NF-kappaB and Erk1/2 signaling pathways in LPS-induced Raw264.7 cells. J Ethnopharmacol 2018;210:95-106. [Crossref] [PubMed]
- Testro AG, Visvanathan K. Toll-like receptors and their role in gastrointestinal disease. J Gastroenterol Hepatol 2009;24:943-54. [Crossref] [PubMed]
- Noh JH, Kim KM, Pandey PR, et al. Loss of RNA-binding protein GRSF1 activates mTOR to elicit a proinflammatory transcriptional program. Nucleic Acids Res 2019;47:2472-86. [Crossref] [PubMed]
- Hunt A. Sepsis: an overview of the signs, symptoms, diagnosis, treatment and pathophysiology. Emerg Nurse 2019;27:32-41. [Crossref] [PubMed]
- Noh JH, Kim KM, Idda ML, et al. GRSF1 suppresses cell senescence. Aging (Albany NY) 2018;10:1856-66. [Crossref] [PubMed]
- Buras JA, Holzmann B, Sitkovsky M. Animal models of sepsis: setting the stage. Nat Rev Drug Discov 2005;4:854-65. [Crossref] [PubMed]
- Huang J, Xuan D, Li X, et al. The value of APACHE II in predicting mortality after paraquat poisoning in Chinese and Korean population: A systematic review and meta-analysis. Medicine (Baltimore) 2017;96:e6838 [Crossref] [PubMed]
- Lambden S, Laterre PF, Levy MM, et al. The SOFA score-development, utility and challenges of accurate assessment in clinical trials. Crit Care 2019;23:374. [Crossref] [PubMed]
(English Language Editor: J. Jones)


