
KCNQ1OT1 lncRNA affects the proliferation, apoptosis, and chemoresistance of small cell lung cancer cells via the JAK2/STAT3 axis
Introduction
Lung cancer, with 1,800,000 new cases worldwide each year, is the most malignant cancer in males and females (1). Small cell lung cancer (SCLC) accounts for 15–20% of lung tumors, and is a high-grade aggressive tumor with rapid growth, early invasiveness, and a tendency to distant metastasis, and has a 5-year survival of approximately 5% (2-4). The National Comprehensive Cancer Network (NCCN) recommends surgery to treat stage I SCLC patients. Although most SCLCs are initially responsive to chemotherapy and radiotherapy, a majority of SCLCs inevitably develop multidrug resistance within 1–2 years (5,6), leading to the low survival of SCLC patients over the past decades. Thus, to explore potential therapies for SCLC, it is essential to delineate the mechanisms underlying the tumor growth, invasion, and chemoresistance of SCLC.
Long noncoding RNAs (lncRNAs), which are >200 nucleotides in length, are clusters of highly conserved evolutionary transcripts without the capacity of protein coding (7,8). There is growing evidence that lncRNAs play vital roles in tumorigenesis and progression, including cell proliferation, differentiation, apoptosis, migration, and invasion. For example, the lncRNA, GAS5, impairs the proliferation and invasion of endometrial carcinoma induced by high glucose by targeting miR-222-3p/p27 (9). The lncRNA, LINC00963, promotes tumorigenesis and radioresistance in breast cancer by sponging miR-324-3p and inducing ACK1 expression (10). In addition, an antisense lncRNA, KCNQ1OT1, which is located at 11p15.5, has attracted extensive interest, as its dysregulation during tumor development accelerates not only cell proliferation, but also chemoresistance. KCNQ1OT1 acts as a competing endogenous RNA (ceRNA) by capturing miR-4458. It enhances osteosarcoma progression by regulating the expression of the cell cycle mediator, cyclin D2 (CCND2) (11). Importantly, knockdown of KCNQ1OT1 improves the sensitivity of osteosarcoma cells to cisplatin by upregulating DNMT1-dedicated Kcnq1 expression (12). However, the roles of KCNQ1OT1 in SCLC have not yet been reported. In the present study, we investigated the function of KCNQ1OT1 in SCLC and found that the lncRNA, KCNQ1OT1, showed higher expression in SCLC chemo-resistant cells than it did in chemo-sensitive cells. The proliferation, cloning ability, migration, and invasion of SCLC cells were hindered and the apoptosis rates increased after KCNQ1OT1 knockdown, suggesting that KCNQ1OT1 promotes tumor growth and chemoresistance in vitro. Further experimentation demonstrated that KCNQ1OT1 could promote the growth and chemoresistance of xenograft SCLC in vivo.
As Janus kinase (JAK) is a protein phosphorylase and phosphorylates signal transducer and activator of transcription (STAT) to bind to DNA, the JAK/STAT pathway is well known to be correlated with cell proliferation and the cell cycle in various malignancies, including gastric cancer (13) and head and neck squamous cell carcinoma (14). Accumulating evidence has shown that lncRNAs interact with JAK2/STAT3, promoting M1 macrophage polarization in children with pneumonia (15) and apoptosis in ovarian cancer (16). Collectively, the above-mentioned studies led us to hypothesize that lncRNAs are involved in the development of SCLC by affecting the JAK2/STAT3 signaling pathway, which may serve as a new therapeutic target for SCLC. In addition to discovered that KCNQ1OT1 inhibition diminishes chemoresistance, cell viability, and invasion, we also found that KCNQ1OT1 affects JAK2/STAT3 signaling.
We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-21-1761).
Methods
Cell lines and cell culture
The human SCLC cell lines, NCI-H69 and NCI-H446, and the multidrug-resistant cell line, NCI-H69AR, were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA), as described in our previous study (17). A drug-resistant subline, H446DDP, was established in our laboratory by culturing H446 cells in cisplatin (DDP; Shandong, China). Cells were maintained in RPMI-1640 medium containing 10% fetal bovine serum (FBS; Gibco, CA, USA) at 37 °C in a humidified atmosphere containing 5% CO2.
Expression profile analysis of lncRNAs
A pair of chemo-resistant and parental SCLC cells was used for expression profiling. RNA extraction and microarray hybridization were performed by Kangchen Company (Shanghai, China) using a human lncRNA microarray (v.3.0; Arraystar, Inc., MD, USA). The data were analyzed by means of GeneSpring 12.6 software (Agilent Technologies, Santa Clara, CA, USA) and the raw signals were log-transformed and normalized using the percentile shift normalization method, with the value set at the 75th percentile. Data processing and statistical analysis for lncRNA data were performed, and heat maps were generated.
Bioinformatic analyses
For KCNQ1OT1 expression analyses, 23 SCLC and 42 normal tissue samples of Affymetrix Human Genome U133 Plus 2.0 Array from the SCLC microarray data set GSE43346 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE43346) were included. Different expression of KCNQ1OT1 was analyzed in the samples to explore the roles in SCLC carcinogenicity.
The interactions between KCNQ1OT1 and its targets were predicted by StarBase 2.0 (http://starbase.sysu.edu.cn/) and the results were cross-referenced with the Pearson correlation analysis to enhance the reliability. To identify functions of KCNQ1OT1 in SCLC, we performed Gene Ontology (GO) function enrichment analysis. In addition, Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.me.jp/kegg) pathway analysis was conducted to annotate the potential functions.
Lentivirus-mediated RNA interference
Two short hairpin RNAs (shRNAs) were targeted on KCNQ1OT1: KCNQ1OT1 sh1#: 5'-GCUCCCAUCUGCACCUUAUTT-3' and KCNQ1OT1 sh2#: 5'-CUUGAAGGGCCACCACUATT-3'. The 2 shRNAs were synthesized and inserted into the lentivirus containing H1 promoter to drive the expression of the shRNA. Lentivirus transduction was performed to downregulate the levels of KCNQ1OT1 (GenePharma, Shanghai, China) in H69AR and H446DDP cells according to the manufacturer’s protocol. Cells were harvested for total RNA extraction and analysis after 2 days.
Quantitative real-time polymerase chain reaction
Total RNA was extracted from cell lines using TRIzol reagent (Invitrogen, Waltham, MA, USA) following the manufacturer’s instructions. The reverse transcription reaction was performed using the PrimeScript RT reagent kit (Tiangen, Beijing, China). The primers to detect the levels of KCNQ1OT1 were as follows: forward, 5'-GCCTGGCCAATCAGAGTACC-3'; reverse, 5'-AGCTGACACGTAGTTC-3'; GAPDH forward, 5'- GAGTCAACGGATTTGGTCGT-3'; and GAPDH reverse, 5'-CATGGGTGGAATCATATTGGA-3'. The levels of expression were calculated with the 2-△△Ct method and normalized to GAPDH.
Cell counting kit-8 assay
The Cell Counting Kit (CCK-8) assay was performed to detect cell viability. Cells (5×103) were cultured in 96-well plates and incubated with CCK8 (Beyotime Institute of Biotechnology, Shanghai, China). The absorbance was measured at 450 nm. In addition, cisplatin (DDP, Shandong, China), adriamycin (ADM, Jiangsu, China), and etoposide (VP-16, Jiangsu, China) cytotoxicity against cells were determined using CCK-8. Cells were incubated in 10 µL of CCK-8 for 2 hours and the absorbance at 450 nm was measured. The half-maximal inhibitory concentration (IC50) of anticancer drugs was calculated according to a cell viability curve.
Flow cytometric analysis
Cells transfected with lentiviral shRNAs were harvested 24 hours after treatment with drugs, resuspended in 400 µL of 1× binding buffer, and then double-stained with 5 µL of annexin V-FITC and 10 µL of the eFluor 780 Kit (eBioscience, San Diego, CA, USA).
Colony formation assay
Transfected H69AR or H446DDP cells were trypsinized and plated in 6-well plates (100 cells/well), which was followed by 2 weeks of incubation. Colonies were fixed and stained. Visible colonies consisting of >50 cells were counted manually.
Cell invasion and migration assay
Invasion assays were performed using 24-well Matrigel invasion chambers (Corning, NY, USA). Cells (3×104) in 200 µL of serum-free medium were plated into the upper chamber. The cells in the bottom chamber were stained with 0.1% crystal violet after 48 hours and then counted. Additionally, a wound healing assay was performed to determine cell migration. The transfected cells were seeded on the 6-well plates. Then, an artificial scratch wound was made using a 100-µL pipette tip. Cells were cultured in serum-free medium for 24 hours, and images were obtained under a light microscope 0 and 24 hours after the wound was made.
Tumor xenograft experiments
All animal experiments were approved by the institutional guidelines of Guangdong Province and Animal Care and Use Committee, and were conducted in accordance with its recommendations and ethical regulations. Male BALB/c nude mice, 3–4 weeks old, were purchased from the Experimental Animal Center of Sun Yat-sen University (Guangzhou, China) and were fed in SPF animal room, and were divided into four groups randomly. The cages and pads were changed every 5 days, and the cages were randomly used after disinfection. The tumor size and body weight of the mice were measured regularly. The total amount of injection in the experimental group and the control group remained the same, as did the solvent. Chemo-resistant H69AR cells (1×107) transfected with lentiviral shRNA or negative control (NC) were subcutaneously injected into the right flanks of BALB/c nude mice. Tumor size was monitored every 3–4 days after tumor formation, and mice were euthanized after 4 weeks. The animals were treated with chemotherapeutics or phosphate-buffered saline (PBS) via intraperitoneal injection (7 mg/kg of etoposide every second day and 3 mg/kg cisplatin at 8-day intervals). Each group had four mice randomly, and there were 16 mice. The allocation was by Weimei Huang; the conduct of the experiment was by Yaru Zhu; the outcome assessment was by Rui Chen; the data analysis was by Yefeng Shen; Eachone did experiment individually.
Western blot
Cells were lysed using RIPA buffer, and protein concentrations were determined by the bicinchoninic acid (BCA) method (BCA Protein Assay Kit; CoWin Biosciences, Taizhou, China). Next, 10 µg of protein were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS‐PAGE), and then transferred to polyvinylidene fluoride (PVDF) membranes. The PVDF membranes were incubated with primary antibodies at 4 °C overnight after incubation in 5% nonfat milk for 1 hour, and incubated with a horseradish peroxidase (HRP)-labeled secondary antibody. After washing again with tris-buffered saline with Tween20 (TBST), immune complexes were detected by chemiluminescence (ECL).
Immunohistochemical staining
The formalin-fixed, paraffin-embedded tissues were sectioned. Sample sections were deparaffinized through a series of xylene baths and alcohol at a high temperature and high pressure for antigen repair. The sections were sliced into a 3% hydrogen peroxide solution and incubated with primary antibody (JAK2, 1:500; Abcam, Cambridge, UK) at 4 °C overnight. Then, the sections were incubated with the secondary antibody [HRP marker, 1:200; Cell Signaling Technology (CST), Danvers, MA, USA]. Immunohistochemical staining was based on the percentage and intensity of the stained cells. The staining intensity ranged from 0 to 3 (0= negative, 1= weak positive, 2= medium strong positive, and 3= strong positive). The H-score was calculated by a semiquantitative assessment of the staining intensity and the percentage of positive cells.
Statistical analysis
The data are presented as the mean ± standard deviation (SD) from 3 or more independent experiments. Differences between groups were analyzed with Student’s t-test and one-way analysis of variance (ANOVA). The two groups were compared using the t test (Student’s t-test).LSD test (homogeneity of variance), Dunnett test and St3 test (heterogeneity of variance) were used for multiple comparisons, P value <0.05 was considered statistically significant.
Results
The expression of the lncRNA, KCNQ1OT1, was linked to SCLC chemoresistance
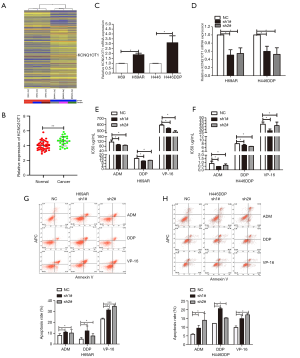
LncRNA expression profiling by microarray analysis was performed for multidrug-resistant (H69AR) and matching sensitive cells (H69). In all, 1,443 lncRNAs (985 upregulated and 458 downregulated), accounting for 4.37% (1,443/33,045) of all lncRNAs, were differentially expressed and were applied to hierarchical cluster analysis. Of note, the lncRNA, KCNQ1OT1, was distinctly upregulated in multidrug-resistant cells compared with sensitive cells (Figure 1A). Using the SCLC Gene Expression Omnibus (GEO) data set, we discovered that KCNQ1OT1 had significantly higher expression in tumor tissues that in normal tissues, indicating that KCNQ1OT1 might be related to SCLC (Figure 1B). Therefore, the expression of KCNQ1OT1 was further analyzed in chemo-resistant versus chemo-sensitive H69 and H446 cells by quantitative polymerase chain reaction (qPCR). KCNQ1OT1 upregulation could be observed in both chemo-resistant SCLC cell lines. In H69AR cells, a 1.9-fold higher expression was observed in comparison to the matching chemo-sensitive H69 cells; meanwhile, in the chemo-resistant H446 cells (H446DDP), the expression was 3.1-fold higher than in the matched sensitive cells (Figure 1C). To explore the role of KCNQ1OT1 in SCLC chemoresistance, we used a lentiviral knock down vector in order to stably silence the KCNQ1OT1 expression. qPCR revealed that the KCNQ1OT1 levels were effectively downregulated by 49% and 45% in H69AR cells and 40% and 47% in H446DDP cells by the two different lentiviral shRNA constructs, respectively (Figure 1D). Subsequently, the effect of the KCNQ1OT1 knockdown on chemoresistance was determined using a viability test under chemotherapeutic pressure. The half-maximal inhibitory concentration (IC50) values of the chemotherapeutics ADM, DDP, and VP-16 were significantly decreased in H69AR and H446DDP cells in response to repression of KCNQ1OT1 (Figure 1E,F; Table S1). Moreover, flow cytometry showed that the apoptosis rates were significantly increased in shKCNQ1OT1-transfected cells after anticancer drug treatment (Figure 1G,H). Thus, the results validated our hypothesis that KCNQ1OT1 was associated with SCLC chemoresistance.
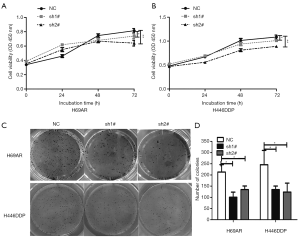
Knockdown of KCNQ1OT1 inhibited SCLC cell viability and cloning ability
Next, we studied the cell viability after suppression of KCNQ1OT1 in H69AR and H446DDP cells and found that there was only moderate, but statistically significant change in cell viability in chemo-resistant H69AR or H446DDP cell lines after KCNQ1OT1 silencing (Figure 2A,B). However, the number of cell colony formation units was significantly decreased by about 50% in KCNQ1OT1-knockdown H69AR and H446DDP cells compared with the NC group (Figure 2C,D). Thus, the results showed that KCNQ1OT1 knockdown markedly inhibited cell viability and cloning ability.
KCNQ1OT1 knockdown hindered SCLC cell migration and invasion and induced apoptosis
Wound-healing experiments revealed that wound closure was less in the shKCNQ1OT1 chemo-resistant cells compared to the NC group (Figure 3A,B,C). Furthermore, Transwell assays demonstrated that knockdown of KCNQ1OT1 more significantly suppressed cell invasion in shKCNQ1OT1-transfected H69AR and H446DDP cells as compared to the NC group (Figure 3D,E). The effects of KCNQ1OT1 on apoptosis of H69AR and H446DDP cells were detected by flow cytometry. The apoptosis rates in shKCNQ1OT1-transfected H69AR and H446DDP cells were much higher than those of the NC group (Figure 3F,G). Therefore, the results revealed that KCNQ1OT1 knockdown hindered SCLC cell migration and invasion, and induced apoptosis.

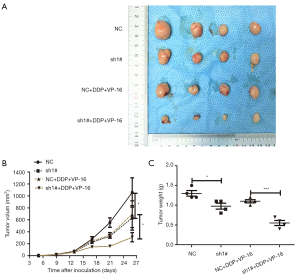
KCNQ1OT1 knockdown suppressed tumor growth and chemoresistance in vivo
To evaluate the tumorigenicity and biological consequences of KCNQ1OT1 in SCLC, we investigated the role of KCNQ1OT1 in tumor growth and chemoresistance in vivo. We implemented a subcutaneous injection of H69AR cells with KCNQ1OT1 knockdown and NC into nude mice (Figure 4A). Compared with the NC group, the KCNQ1OT1-knockdown group showed repressed tumor volume growth. The tumor size in the group treated with shKCNQ1OT1 + anticancer drugs was markedly smaller than that in the NC + anticancer drug group (Figure 4B), and the difference in the weight of the tumors after treatments also displayed a similar tendency (Figure 4C). Taken together, knockdown of KCNQ1OT1 suppressed tumor growth and decreased chemoresistance to anticancer drugs.
KCNQ1OT1 reduced SCLC tumor growth and chemoresistance via the JAK2/STAT3 signaling pathway
In order to explore the potential target messenger RNA (mRNA) and pathways, 96 altered lncRNA‐related mRNAs were determined by StarBase. Furthermore, we analyzed GO enrichment and KEGG pathway using DAVID (Database for Annotation, Visualization and Integrated Discovery; Figure 5A) to identify the potential functions of target mRNA. The top 10 GO terms were poly A RNA binding, RNA binding, nucleotide binding, mRNA splicing, mRNA processing, nucleoplasm, gene expression, nucleus, intracellular ribonucleoprotein complex, and nucleic acid binding (Figure 5B). Meanwhile, KEGG pathway analysis revealed that altered mRNAs were enriched in spliceosome, RNA transport, mRNA surveillance pathway, ribosome biogenesis in eukaryotes and amyotrophic lateral sclerosis (Figure S1).
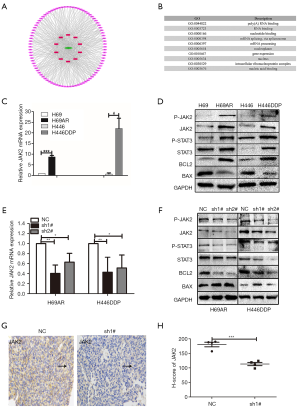
The relevant and literature and results of GO and KEGG pathways, led us to believe that JAK2/STAT3 pathway might be linked to KCNQ1OT1 in SCLC chemoresistance (18). To test our hypothesis that JAK2/STAT3 signaling pathway is involved in SCLC chemoresistance, we studied JAK2/STAT pathway activation for the potential regulatory mechanisms. We focused on the phosphorylation of JAK2 and STAT3 as JAK2/STAT3 pathway-related markers, including P-JAK2, JAK2, P-STAT3, and STAT3. Furthermore, B-cell lymphoma 2 (BCL2) and BCL-2-associated X protein (BAX) expression was studied due to key survival factors downstream of oncogenic JAK2 (19). Here, we found that JAK2 significantly differed in the chemo-resistant versus sensitive cells (Figure 5C,D, Figure S2). In elucidating the interaction between KCNQ1OT1 and JAK2/STAT3, we found that mRNA JAK2 was deregulated after KCNQ1OT1 knockdown (Figure 5E). After KCNQ1OT1 knockdown, the expression of pathway‐related marker proteins (P-JAK2, JAK2, P-STAT3, STAT3, and BCL2) was decreased, while BAX expression was elevated (Figure 5F, Figure S2), according to the results of western blot in H69AR and H446DDP cells. To demonstrate the expression of JAK2 in subcutaneous tumors of nude mice, immunostaining was applied. The results, according to the JAK2 H-score, revealed that JAK2 expression was significantly lower in shKCNQ1OT1-transducted xenograft tumor tissues compared with tumors derived from SCLC cells transduced with NC (Figure 5G,H). The results suggested that KCNQ1OT1 promoted activation of the JAK2/STAT3 signaling pathway.
Discussion
SCLC is a common malignancy with a poor prognosis and has thus imposed an enormous burden on public health. In recent decades, no new therapies or insights into the satisfactory treatment of SCLC patients have been developed. In a search for a potential therapy target for SCLC, lncRNAs have considerable great attention as oncogenes or tumor suppressors due to their ability to regulate cell proliferation, migration, invasion, and chemoresistance. Numerous studies have illustrated that a newly identified lncRNA, KCNQ1OT1, facilitates the progression and metastasis of non-small cell lung, breast, bladder, and colorectal cancer (20-23), while inducing chemoresistance of hepatocellular carcinoma and acute myeloid leukemia (24,25). In the current study, microarray analysis of lncRNA expression profiles indicated that KCNQ1OT1 is involved in SCLC chemoresistance. To better understand how KCNQ1OT1 affects chemoresistance, we used two efficient lentiviral shRNAs to downregulate the levels of KCNQ1OT1, exploring the function of KCNQ1OT1. After downregulating the expression of KCNQ1OT1, the proliferation, cloning ability, migration, and invasion of SCLC cells were inhibited and the apoptosis rates were increased, suggesting that KCNQ1OT1 promoted tumor growth and chemoresistance in vitro. Further, KCNQ1OT1 promoted the growth and chemoresistance of xenograft SCLC in vivo. Moreover, KCNQ1OT1 knockdown might inhibit the activation of the JAK2/STAT3 signaling pathway.
Currently, lncRNA KCNQ1OT1 has been demonstrated to function in numerous cancers, including tongue cancer (26), colorectal cancer (22), non-small cell lung cancer (20), and bladder cancer (23). In colorectal cancer, ZEB1-induced upregulation of KCNQ1OT1 improved the proliferation, migration and epithelial-mesenchymal transition (EMT) formation of colorectal cancer cells via regulation of the miR-217/ZEB1 axis (27). Similarly, we found that KCNQ1OT1 promoted the proliferation, cloning, migration, and invasion of cancer cells. In addition, the knockdown of KCNQ1OT1 has been reported to significantly induce the apoptosis of osteosarcoma cells (28) and non-small cell lung cancer cells (20), while the silencing of KCNQ1OT1 was shown to impede methotrexate-resistant colorectal cancer cell tumor growth in nude mice (22). In vivo, we discovered that the downregulation of KCNQ1OT1 repressed the volume growth and reduced the weight of the tumors, indicating that KCNQ1OT1 may acts as a carcinogenic and chemo-resistant biomarker for SCLC.
After predicting potential target mRNAs by bioinformatic tools, we identified 96 altered lncRNA‐related mRNAs of KCNQ1OT1. Furthermore, we conducted GO and KEGG pathways to determine the possible biological functions and molecular regulatory mechanisms of target mRNAs of KCNQ1OT1. The GO terms indicated that most altered mRNAs were enriched in poly A RNA binding, RNA binding, nucleotide binding, mRNA splicing, and mRNA processing. However, KEGG pathways suggested the interaction with several cancer-related pathways, including viral carcinogenesis, microRNAs and transcriptional misregulation in cancer.
Recently, it has been demonstrated that activation of the JAK2/STAT3 signaling pathway results in tumorigenesis and progression in various cancers, including ovarian (29), bladder (30), colorectal (31), and cervical cancers (32). Moreover, lncRNA has been correlated with the tumor growth factor beta (TGF-β) pathway and the interleukin (IL)-6/JAK2/STAT3 pathway, through which it can promote the proliferation and migration of advanced chronic myeloid leukemia (33) and non-small cell lung cancer (34). Using the evidence and results of GO and KEGG, we showed that KCNQ1OT1 knockdown induced drastic changes in JAK2/STAT3 pathway‐related markers and activated the pathway. According to previous studies, the lncRNA, TUG1, contributes to the proliferation and migration of hepatocellular carcinoma cells in vivo by upregulating miR-144 via inactivation of the JAK2/STAT3 pathway (35). Moreover, the lncRNA, HOTAIR, was found to promote multiple myeloma cell viability and increase chemoresistance to dexamethasone via the JAK2/STAT3 signaling pathway (36). We are of the opinion that KCNQ1OT1 competitively binds to microRNAs and regulates the downstream oncogenes or signaling pathways. There is also the possibility that KCNQ1OT1 promotes DDP resistance via DNA repair pathway, in similar fashion to lncRNA DDSR1 in homologous recombination (37). To further this research, we are conducting corollary experiments to better understand how KCNQ1OT1 activates the JAK2/STAT3 signaling pathway in SCLC.
Conclusions
Our study is the first to report that the lncRNA, KCNQ1OT1, promotes cell proliferation, invasion, and chemoresistance, and suppresses apoptosis via the JAK2/STAT3 signaling pathway in SCLC. Furthermore, our results suggest that KCNQ1OT1 may serve as a potential therapeutic target in SCLC.
Acknowledgments
Funding: YS is supported by grants from the Oversea Study Program of Guangzhou Elite Project. Further funding for this study was provided by the Natural Science Foundation of Guangdong Province (No. 2016A030313137).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-1761
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-1761
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-1761). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All animal experiments were approved by the institutional guidelines of Guangdong Province and Animal Care and Use Committee, and were conducted in accordance with its recommendations and ethical regulations.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Govindan R, Page N, Morgensztern D, et al. Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: analysis of the surveillance, epidemiologic, and end results database. J Clin Oncol 2006;24:4539-44. [Crossref] [PubMed]
- Bayman NA, Sheikh H, Kularatne B, et al. Radiotherapy for small-cell lung cancer-Where are we heading? Lung Cancer 2009;63:307-14. [Crossref] [PubMed]
- Byers LA, Rudin CM. Small cell lung cancer: where do we go from here? Cancer 2015;121:664-72. [Crossref] [PubMed]
- Semenova EA, Nagel R, Berns A. Origins, genetic landscape, and emerging therapies of small cell lung cancer. Genes Dev 2015;29:1447-62. [Crossref] [PubMed]
- Kalemkerian GP, Schneider BJ. Advances in Small Cell Lung Cancer. Hematol Oncol Clin North Am 2017;31:143-56. [Crossref] [PubMed]
- Harrow J, Frankish A, Gonzalez JM, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 2012;22:1760-74. [Crossref] [PubMed]
- Zhao Y, Li H, Fang S, et al. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res 2016;44:D203-8. [Crossref] [PubMed]
- Li Z, Yu Z, Meng X, et al. Long noncoding RNA GAS5 impairs the proliferation and invasion of endometrial carcinoma induced by high glucose via targeting miR-222-3p/p27. Am J Transl Res 2019;11:2413-21. [PubMed]
- Zhang N, Zeng X, Sun C, et al. LncRNA LINC00963 Promotes Tumorigenesis and Radioresistance in Breast Cancer by Sponging miR-324-3p and Inducing ACK1 Expression. Mol Ther Nucleic Acids 2019;18:871-81. [Crossref] [PubMed]
- Wang M, Wang Z, Zhu X, et al. LncRNA KCNQ1OT1 acting as a ceRNA for miR-4458 enhances osteosarcoma progression by regulating CCND2 expression. In Vitro Cell Dev Biol Anim 2019;55:694-702. [Crossref] [PubMed]
- Qi X, Yu XJ, Wang XM, et al. Knockdown of KCNQ1OT1 Suppresses Cell Invasion and Sensitizes Osteosarcoma Cells to CDDP by Upregulating DNMT1-Mediated Kcnq1 Expression. Mol Ther Nucleic Acids 2019;17:804-18. [Crossref] [PubMed]
- Li X, Na H, Xu L, et al. DC-SIGN mediates gastric cancer progression by regulating the JAK2/STAT3 signaling pathway and affecting LncRNA RP11-181G12.2 expression. Biomed Pharmacother 2020;121:109644 [Crossref] [PubMed]
- Jin S, Yang X, Li J, et al. p53-targeted lincRNA-p21 acts as a tumor suppressor by inhibiting JAK2/STAT3 signaling pathways in head and neck squamous cell carcinoma. Mol Cancer 2019;18:38. [Crossref] [PubMed]
- Chi X, Ding B, Zhang L, et al. lncRNA GAS5 promotes M1 macrophage polarization via miR-455-5p/SOCS3 pathway in childhood pneumonia. J Cell Physiol 2019;234:13242-51. [Crossref] [PubMed]
- Sun Q, Li Q, Xie F. LncRNA-MALAT1 regulates proliferation and apoptosis of ovarian cancer cells by targeting miR-503-5p. Onco Targets Ther 2019;12:6297-307. [Crossref] [PubMed]
- Wang Q, Zeng F, Sun Y, et al. Etk Interaction with PFKFB4 Modulates Chemoresistance of Small-cell Lung Cancer by Regulating Autophagy. Clin Cancer Res 2018;24:950-62. [Crossref] [PubMed]
- Xue YH, Ge YQ. Construction of lncRNA regulatory networks reveal the key lncRNAs associated with Pituitary adenomas progression. Math Biosci Eng 2020;17:2138-49. [Crossref] [PubMed]
- Waibel M, Solomon VS, Knight DA, et al. Combined targeting of JAK2 and Bcl-2/Bcl-xL to cure mutant JAK2-driven malignancies and overcome acquired resistance to JAK2 inhibitors. Cell Rep 2013;5:1047-59. [Crossref] [PubMed]
- Kang Y, Jia Y, Wang Q, et al. Long Noncoding RNA KCNQ1OT1 Promotes the Progression of Non-Small Cell Lung Cancer via Regulating miR-204-5p/ATG3 Axis. Onco Targets Ther 2019;12:10787-97. [Crossref] [PubMed]
- Feng W, Wang C, Liang C, et al. The Dysregulated Expression of KCNQ1OT1 and Its Interaction with Downstream Factors miR-145/CCNE2 in Breast Cancer Cells. Cell Physiol Biochem 2018;49(:432-46.
- Xian D, Zhao Y. LncRNA KCNQ1OT1 enhanced the methotrexate resistance of colorectal cancer cells by regulating miR-760/PPP1R1B via the cAMP signalling pathway. J Cell Mol Med 2019;23:3808-23. [Crossref] [PubMed]
- Wang J, Zhang H, Situ J, et al. KCNQ1OT1 aggravates cell proliferation and migration in bladder cancer through modulating miR-145-5p/PCBP2 axis. Cancer Cell Int 2019;19:325. [Crossref] [PubMed]
- Hu H, Yang L, Li L, et al. Long non-coding RNA KCNQ1OT1 modulates oxaliplatin resistance in hepatocellular carcinoma through miR-7-5p/ABCC1 axis. Biochem Biophys Res Commun 2018;503:2400-6. [Crossref] [PubMed]
- Sun H, Sun Y, Chen Q, et al. LncRNA KCNQ1OT1 contributes to the progression and chemoresistance in acute myeloid leukemia by modulating Tspan3 through suppressing miR-193a-3p. Life Sci 2020;241:117161 [Crossref] [PubMed]
- Zhang S, Ma H, Zhang D, et al. LncRNA KCNQ1OT1 regulates proliferation and cisplatin resistance in tongue cancer via miR-211-5p mediated Ezrin/Fak/Src signaling. Cell Death Dis 2018;9:742. [Crossref] [PubMed]
- Bian Y, Gao G, Zhang Q, et al. KCNQ1OT1/miR-217/ZEB1 feedback loop facilitates cell migration and epithelial-mesenchymal transition in colorectal cancer. Cancer Biol Ther 2019;20:886-96. [Crossref] [PubMed]
- Shen Y, Xu J, Pan X, et al. LncRNA KCNQ1OT1 sponges miR-34c-5p to promote osteosarcoma growth via ALDOA enhanced aerobic glycolysis. Cell Death Dis 2020;11:278. [Crossref] [PubMed]
- Fogg KC, Olson WR, Miller JN, et al. Alternatively activated macrophage-derived secretome stimulates ovarian cancer spheroid spreading through a JAK2/STAT3 pathway. Cancer Lett 2019;458:92-101. [Crossref] [PubMed]
- Chen Z, Du Y, Liu X, et al. EZH2 inhibition suppresses bladder cancer cell growth and metastasis via the JAK2/STAT3 signaling pathway. Oncol Lett 2019;18:907-15. [Crossref] [PubMed]
- He T, Cui J, Wu Y, et al. Knockdown of TRIM66 inhibits cell proliferation, migration and invasion in colorectal cancer through JAK2/STAT3 pathway. Life Sci 2019;235:116799 [Crossref] [PubMed]
- Zeng YT, Liu XF, Yang WT, et al. REX1 promotes EMT-induced cell metastasis by activating the JAK2/STAT3-signaling pathway by targeting SOCS1 in cervical cancer. Oncogene 2019;38:6940-57. [Crossref] [PubMed]
- Li ZY, Yang L, Liu XJ, et al. The Long Noncoding RNA MEG3 and its Target miR-147 Regulate JAK/STAT Pathway in Advanced Chronic Myeloid Leukemia. EBioMedicine 2018;34:61-75. [Crossref] [PubMed]
- Shi J, Li J, Yang S, et al. LncRNA SNHG3 is activated by E2F1 and promotes proliferation and migration of non-small-cell lung cancer cells through activating TGF-beta pathway and IL-6/JAK2/STAT3 pathway. J Cell Physiol 2020;235:2891-900. [Crossref] [PubMed]
- Lv J, Kong Y, Gao Z, et al. LncRNA TUG1 interacting with miR-144 contributes to proliferation, migration and tumorigenesis through activating the JAK2/STAT3 pathway in hepatocellular carcinoma. Int J Biochem Cell Biol 2018;101:19-28. [Crossref] [PubMed]
- Guan R, Wang W, Fu B, et al. Increased lncRNA HOTAIR expression promotes the chemoresistance of multiple myeloma to dexamethasone by regulating cell viability and apoptosis by mediating the JAK2/STAT3 signaling pathway. Mol Med Rep 2019;20:3917-23. [Crossref] [PubMed]
- Sharma V, Khurana S, Kubben N, et al. A BRCA1-interacting lncRNA regulates homologous recombination. EMBO Rep 2015;16:1520-34. [Crossref] [PubMed]
(English Language Editor: J. Gray)


