
Isolation of exosomes from serum of patients with lung cancer: a comparison of the ultra-high speed centrifugation and precipitation methods
Introduction
Exosomes are nano capsules secreted by cells through endogenous pathways. They have a cup-shaped or concave hemispherical structure, with a particle size distribution of 30–200 nm (1). Exosomes can be found in a range of bodily fluids including blood, urine, ascites, saliva, amniotic fluid, milk, semen, and cerebrospinal fluid. These extracellular vesicles (EVs) contain a variety of lipids, proteins, and nuclei acids, which act as intracellular biological signals to adjacent or distal cells for uptake by target cells (2). Therefore, exosomes are widely involved in physiological and pathological processes and play an important role in cell communication. Furthermore, exosomes have been recognized as the cause of many diseases and as such, may act as potential biomarkers or therapeutic targets (3).
The classical classification of EVs includes apoptotic bodies, microvesicles, and exosomes (4,5), all of which act via different mechanisms, have different particle sizes, and varying content. Apoptotic bodies and microvesicles are formed by direct budding of cells, but apoptotic bodies can only bud from apoptotic cells. However, complicated endogenous pathways are involved in the formation of exosomes (6-8). First, the cell membrane invaginates to form early endosomes. The membrane of the early endosome, forming intracavitary vesicles in the early endosome, and then matured into the late endosome. In the late stage, the internal body membrane continues to invaginate and the vesicles in the cavity continue to accumulate, thus forming multi-vesicular bodies (MVBs). The multi-vesicular membrane fuses with the cell membrane, and the inner vesicle, that is, the exosome, is then expelled. Despite the different properties of the exosomes at each stage of formation, they cannot be effectively separated due to technical limitations. In fact, the lack of an efficient isolation method is currently a major obstacles in the study of exosomes. This present investigation compared two methods of extracting exosomes from the serum of lung cancer patients, namely, the precipitation kit method and the classical ultra-high speed centrifugation method. Identification and optimization of an efficient extraction method is crucial for future research regarding exosomes.
We present the following article in accordance with the MADR reporting checklist (available at http://dx.doi.org/10.21037/atm-21-2075).
Methods
Sample preparation
Serum samples were collected from a patient with primary lung cancer who received surgical treatment and was confirmed by pathological examination in Renji Hospital in December 2018. The patient voluntarily participated in this research project and signed an informed consent form (approved by Renji Hospital Ethics Committees: 2018-160). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Peripheral venous blood (15 mL) was collected from the patient in a serum separation test tube (without heparin). The sample was kept at room temperature for 30 minutes prior to centrifugation at 2,000 g for 15 minutes at 4 °C. The upper pale-yellow layer of serum was collected and divided into 2 centrifuge tubes each with 3 to 5 mL serum. Samples were kept on ice as much as possible and stored at −80 °C. The above process should not exceed 6 hours.
Instruments and reagents
The Avanti JXN-30/26 Intelligent High-Efficiency Centrifuge and the associated ultracentrifuge tube, Prescot-Exokit, were purchased from a company (Umibio Science and Technology Group, Shanghai, China). The following equipment was used: a biological transmission electron microscope (Tecnai G2 Spirit Biotin/*Tecnai G2 Spirit Biotin), a FEI Talos F200C 200 kV electron microscope, a carbon support membrane for the Mesoscope, and a Malvern NanoSight NS300.
Extraction of exosomes from serum by ultra-high speed centrifugation
Serum samples were centrifuged at 2,000 g for 5 minutes at 4 °C. The supernatant was transferred to a new tube and centrifuged at 3,500 g for 15–20 minutes at 4 °C. The supernatant was again transferred to a new tube and centrifuged at 10,000 g for 30–40 minutes at 4 °C. The supernatant was then transferred to an ultra-high speed centrifuge tube and centrifuged at 120,000 g for 140 minutes at 4 °C in an ultra-high speed centrifuge. The supernatant was discarded and the precipitate was resuspended in 1× phosphate buffered saline (PBS) and passed through a 0.22 µm sterile filter. The supernatant was again collected and centrifuged at 120,000 g for 70 minutes at 4 °C. The supernatant was then carefully discarded. The small amount of liquid at the bottom of the centrifuge tube containing the serum exosomes was resuspended in 100 mL 1× PBS solution and stored at 4 or −80 °C.
Extraction of exosomes from serum by the precipitation method
For the extraction of serum exosomes via precipitation, the Prekit-Exo kit ((Umibio Science and Technology Group, Shanghai, China) was used according to the manufacturer’s instructions. Serum samples were centrifuged at 3,000 g for 15–20 minutes at 4 °C. The supernatant was collected and filtered through a 0.45 µm filter. The filtered supernatant was then incubated with the reagent in the PreKit-Exo kit (Vsample liquid: Vkit =1/4) at 4 °C overnight. Samples were then centrifuged at 1,500 g for 30–40 minutes at 4 °C. The resultant supernatant was discarded and the precipitate was centrifuged again at 1,500 g for 5–10 minutes at 4 °C to fully remove the supernatant. The remaining precipitate containing the exosomes was washed with 100 mL 1× PBS and centrifuged for at 1,500 g for 5 minutes. The exosome pellet was then resuspended in PBS and stored at 4 or −80 °C.
Biological transmission electron microscopy of serum exosomes
The exosome suspension (10 mL) was transferred as a droplet into the copper mesh grid of the electron microscope. The drop was left on the copper mesh for more than 3 minutes. After negative staining with 2% uranium diacetate solution for 1 minute, the excess solution was dabbed with a piece of filter paper and the samples were air dried at room temperature. The copper mesh samples were observed and photographed under a 120 kv biological transmission electron microscope.
Cryo-electron microscopy and image processing
The cryo-electron microscopy was performed in the Center of Cryo-Electron Microscopy Zhejiang University. For each glow discharge porous carbon grid (Quantifile CuR 1.2/1.3), a 3.5 µL sample with a particle concentration of about 1 µM was used. The mesh was then sucked dry with Whatman 55 mm filter paper for 1.5 seconds, quickly frozen in liquid nitrogen, and cooled in FEI Vitrobot Mark IV. The grid was transferred to an FEI Talos electron microscope running at an accelerating voltage of 200 kV and equipped with a Gatan K2 Summit direct electronic counting camera. The micrograph was recorded in super-resolution mode using the semi-automatic low-dose acquisition program UCSF- image 4, with a nominal magnification of 22,500×, which is equivalent to the pixel size of 1.31 Å at the sample level. The total exposure time of each image is 8 seconds, which is divided into 32 sub-frames.
Analysis of particle size distribution and concentration of exosomes using the nanolayers particle size tracking meter
Serum exosome samples (10 mL) were dilute to 1,000 mL with deionized water. Samples were loaded with a 1 mL syringe and the concentration and size distribution of exosomes were measure using the nanolaser particle size tracking meter.
Statistical analysis
SPSS 24.0 software was used for statistical analysis. The experimental data were expressed as mean ± standard deviation (). The independent sample t-test and one-way analysis of variance (ANOVA) were applied, and the homogeneity of variance test was performed at the same time. When the variance was homogeneous, the LSD statistical method was used for comparisons between groups. P<0.05 indicated a significant difference.
Results
Transmission electron microscopy and cryo-electron microscopy
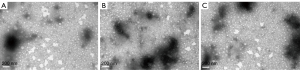
Biotransmission electron microscopy was used to detect the morphology of exosomes extracted from the serum of lung cancer patients. Exosomes obtained by ultra-high speed centrifugation showed obvious cup-shaped or concave hemispherical structure, and some were clustered together (Figure 1). However, the exosomes extracted by the precipitation kit method did not show an obvious structure and impurities could be observed (Figure 2).
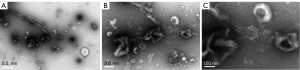
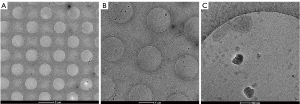
Cryo-electron microscopy showed that exosomes obtained by ultra-high speed centrifugation had obvious circular morphology or Whistle shape structure, as showed in Figure 3.
Nanoparticle tracking analysis (NTA)
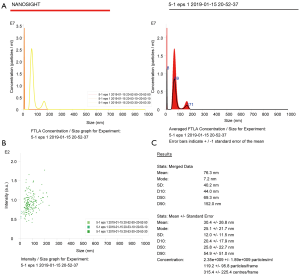
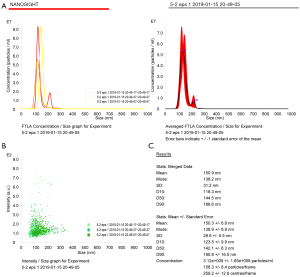
The particle size distribution and concentration of exosomes were analyzed by using a nanolaser particle size tracking meter. Take the sample 3 times at a time.
The average diameter of exosomes obtained by ultra-high speed centrifugation was smaller (30.4±26.8 nm) (Figure 4) than that of exosomes extracted using the precipitation kit method (150.3±6.8 nm) (Figure 5). It is possible that the precipitant in the precipitation kit was extracted together with the exosomes to cause the increased particle size. Furthermore, the particle size of exosomes extracted by the precipitation kit method was within a relatively narrow range (Figure 5), which may be due to the coating effect of the precipitation reagent covering up the differences in the particle sizes of the exosomes themselves.
Discussion
First reported in 1981, exosomes are vesicles produced from cells that can be found in body fluids or cell supernatants (9-12). In recent years, there has been much interest in exosomes due to its role in cell protection, intercellular communication, and disease diagnosis and treatment (13). However, the efficient isolation and purification of exosomes from various sources, especially from the serum, are preliminary problems that need to be resolved (14-16). At present, methods for the extraction and purification of exosomes include ultracentrifugation, membrane separation based on particle size, magnetic bead immunocapture, and precipitation kits. However, each method has its advantages and disadvantages. Many exosome isolation products have come onto the market, however, these products generally have not been standardized and thus it is important to examine the quality of the resultant isolated exosomes (17-22). In this investigation, exosomes isolated and purified from the serum of lung cancer patients by ultracentrifugation and a precipitation kit were compared. The exosomes extracted by ultra-high speed centrifugation were abundant and showed good shape, with cup-shaped or concave hemispherical structure typical of exosomes. The background was clean and exosomes were observed to gather together. Moreover, the exosomes showed a relatively uniform particle size distribution with a small particle size of 30.4±26.8 nm and a NTA concentration of 2.35×109 cells/mL. In contrast, the particle size of the exosomes extracted by the precipitation kit method was larger (150.3±6.8 nm) and within a relatively narrow range. This was likely due to the coating effect of the precipitation reagent masking the differences in particle sizes of the exosomes themselves. Although the classical ultra-high speed centrifugation method requires the use of an ultra-high speed centrifuge, the quality of exosomes extracted by this method was superior to that obtained with the precipitation kit method. Furthermore, the precipitation kit was expensive and the separation purity was of concern. It is crucial that researchers pay attention to the quality and efficacy of the extraction method so as to produce good quality exosomes for further research regarding their properties and functions.
Acknowledgments
The authors wish to thank Dr. Xinqiu Guo (Shanghai Jiaotong University), Dr. Shihua Yao, and the patients involved for their assistance in this study. The authors also thank the help from Center of Cryo-Electron Microscopy, Zhejiang University.
Funding: This work was financially supported by the National Foundational Basic Research Project of China (2017YFA0205304 and 2015CB931802), National Natural Scientific Foundation (81803094 and 81671782), Shanghai Municipal Commission of Economy and Information Technology Fund (No. XC-ZXSJ-02-2016-05), and the Medical Engineering Cross Project of Shanghai Jiaotong University (YG2016ZD10 and YG2017ZD05).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-2075
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-2075
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-2075). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics board of Renji Hospital (No.: 2018-160) and informed consent was taken from all individual participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Peinado H, Alečković M, Lavotshkin S, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med 2012;18:883-91. [Crossref] [PubMed]
- Yang X, Gu Y, Fu X, et al. Research Progress of Exosomes in Lung Cancer. Medical Recapitulate 2018. Available online: http://en.cnki.com.cn/Article_en/CJFDTotal-YXZS201819014.htm
- Huang Y, Zheng L. Attach importance to the experimental diagnostic value of exosomes. J Lab Med 2015;38:724-6.
- Huang Y, Tang Y, Qin S, et al. Comparison of RNA Extraction Methods of Exosomes and Exosomes in serum. J Lab Med 2016;39:427-32.
- Iero M, Valenti R, Huber V, et al. Tumour-released exosomes and their implications in cancer immunity. Cell Death Differ 2008;15:80-8. [Crossref] [PubMed]
- Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654-9. [Crossref] [PubMed]
- Hao S, Ye Z, Li F, et al. Epigenetic transfer of metastatic activity by uptake of highly metastatic B16 melanoma cell-released exosomes. Exp Oncol 2006;28:126-31. [PubMed]
- EL Andaloussi S. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov 2013;12:347-57. [Crossref] [PubMed]
- Yang T, Martin P, Fogarty B, et al. Exosome delivered anticancer drugs across the blood-brain barrier for brain cancer therapy in Danio rerio. Pharm Res 2015;32:2003-14. [Crossref] [PubMed]
- Syn N, Wang L, Sethi G, et al. Exosome-Mediated Metastasis: From Epithelial-Mesenchymal Transition to Escape from Immunosurveillance. Trends Pharmacol Sci 2016;37:606-17. [Crossref] [PubMed]
- Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics 2010;73:1907-20. [Crossref] [PubMed]
- van Niel G, Porto-Carreiro I, Simoes S, et al. Exosomes: a common pathway for a specialized function. J Biochem 2006;140:13-21. [Crossref] [PubMed]
- Sullivan R, Saez F, Girouard J, et al. Role of exosomes in sperm maturation during the transit along the male reproductive tract. Blood Cells Mol Dis 2005;35:1-10. [Crossref] [PubMed]
- Xu CF, Yang ZL, Hua YT, et al. The role of exosomes in drug resistance information transmission of breast cancer in vitro. Journal of Xi'an Jiaotong University 2017;38:193-8.
- Whiteside TL. Exosomes carrying immunoinhibitory proteins and their role in cancer. Clin Exp Immunol 2017;189:259-67. [Crossref] [PubMed]
- Zhang N, Nan A, Chen L, et al. Circular RNA circSATB2 promotes progression of non-small cell lung cancer cells. Mol Cancer 2020;19:101. [Crossref] [PubMed]
- Simpson RJ, Jensen SS, Lim JW. Proteomic profiling of exosomes: current perspectives. Proteomics 2008;8:4083-99. [Crossref] [PubMed]
- Li L, Piontek KB, Kumbhari V, I, et al. Isolation and Profiling of MicroRNA-containing Exosomes from Human Bile. J Vis Exp 2016;54036. [Crossref] [PubMed]
- Johnstone RM, Adam M, Hammond JR, et al. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem 1987;262:9412-20. [Crossref] [PubMed]
- Cheng H. Research Progress of Exosomes in Cardiovascular Diseases. Adv Clin Med 2019;09:42-50. [Crossref]
- Zhao L, Yu J, Wang J, et al. Isolation and Identification of miRNAs in exosomes derived from serum of colon cancer patients. J Cancer 2017;8:1145-52. [Crossref] [PubMed]
- Alipoor SD, Mortaz E, Varahram M, et al. The Potential Biomarkers and Immunological Effects of Tumor-Derived Exosomes in Lung Cancer. Front Immunol 2018;9:819. [Crossref] [PubMed]
(English Language Editor: J. Teoh)


