
Identification of an IFN-β-associated gene signature for the prediction of overall survival among glioblastoma patients
Introduction
Brain glioblastoma multiforme (GBM) is the most common primary malignant intracranial tumor (50%), and is associated with high morbidity and mortality in both adults and children (1-4). A histopathological from low-grade to high-grade transformation is associated with poor overall survival (5). Currently, surgery, radiation, and chemotherapy are the main treatment modalities of GBM. Chemotherapy is a critical process in the postsurgical treatment of GBM (6-8). Alkylating agents, such as temozolomide, remain the standard of care in GBM chemotherapy, but response remains poor (9).
DNA repair protein, O6-methylguanine-DNA methyltransferase (MGMT), plays an essential role in cellular resistance to alkylating agents (10). Clinically, chemoresistance occurs frequently in patients with GBM that exhibit an aberrant activation of MGMT. β-interferon (IFN-β) can act as a drug sensitizer, enhancing toxicity against various neoplasias, and is widely used in combination with other antitumor agents, such as nitrosoureas (11-13). IFN-β sensitizes glioma cells that harbor the unmethylated MGMT promoter and are resistant to temozolomide (11,14,15). Likewise, IFN-β induces loss of spherogenicity and overcomes therapy resistance of glioblastoma stem cells (16). Nevertheless, the specific mechanisms and molecules associated with this phenomenon have not yet been completely elucidated. Therefore, the main objective of the study was to elucidate the molecular mechanisms responsible for the beneficial effect of IFNβ in GBM.
So far, studies have focused mainly on one gene is related with the other. In the present study, we firstly explored and analyzed all differentially expressed IFN-β-associated genes [gene set enrichment analysis (GSEA) M2567] and IFN-β-associated lncRNAs by systematic bioinformatics analysis. In total, 596 GBM patients were included in The Cancer Genome Atlas (TCGA) GBM to construct the prognostic model. Univariate Cox regression model found 5 survival-related genes. Lasso-penalized Cox analysis then identified 5 genes to construct the prognostic model. Using the methodology previously described, the result is validated on the Gene Expression Omnibus (GEO) datasets (GSE83300). We found that a 4 IFN-β-associated gene [peroxiredoxin 1 (PRDX1), Sec61 subunit beta (SEC61B), X-ray repair cross-complementing 5 (XRCC5), and Bcl-2-like protein 2 (BCL2L2)] signature was a robust marker of seizure prognosis in patients with GBM. Using data from the Chinese Glioma Genome Atlas (CGGA), we found that 4 IFN-β-associated genes were independent biomarkers of prognosis. Pathway enrichment analysis results demonstrated that several modules are enriched in GBM-related pathways.
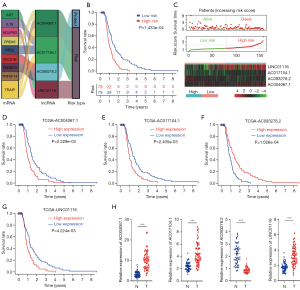
Long non-coding RNA (lncRNA) has been demonstrated to play an important role in human diseases (17), especially in GBM. LncRNA AC003092.1 regulates tissue factor pathway inhibitor-2 (TFPI-2) expression through the competing endogenous RNA mechanism, and lncRNA SOX2OT (SOX2 overlapping transcript) interacts with RNA-binding proteins to promote the expression level of SOX2, which is involved in glioma chemotherapy (18,19). However, the relevance between IFN-β and lncRNA has not been fully elucidated in GBM. Pearson correlation was applied to calculate the correlation between lncRNAs and IFN-β-associated genes. An lncRNA with a correlation coefficient |R2|>0.3 and P<0.05 was considered to be an IFN-β-associated lncRNA. Univariate and multivariate Cox regressions were used for the survival analysis, which indicated that AC093278.2, AC004067.1, LINC01116, and AC017104.1 were independent prognostic factors for the overall survival of GBM patients.
The findings of the present study may lay the foundation for future studies investigating GBM. We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/atm-21-1986).
Methods
Clinical specimens and data collection
Fifty human glioma tissue samples and 50 normal brain tissues were obtained from the Jiangxi Cancer Hospital of Nanchang University, China. Samples were frozen in liquid nitrogen after surgical resection. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The present study was approved by the Ethics Committee of the Jiangxi Cancer Hospital of Nanchang University. Informed consent was obtained from patients or guardians.
Messenger RNA (mRNA) expression profiles and clinical data were obtained from TCGA GBM (https://cancergenome.nih.gov/), the GEO (GSE83300) database (https://www.ncbi.nlm.nih.gov/geo/), and the CGGA (http://www.cgga.org.cn/). The IFN-β-associated gene set (M2567) was obtained by GSEA using the gene set database (http://www.gsea-msigdb.org/). All data were analyzed using R software (version 4.0.2) (http://www.r-project.org).
Identification of differentially expressed genes in TCGA GBM
The limma package was used to screen the differentially expressed genes of interest with R software version 4.0.2. The expression pattern of the 120 IFN-β-associated genes was then investigated in TCGA. Genes were selected as consistently altered IFN-β-associated genes for subsequent prognostic analysis if they demonstrated a consistent expression pattern in TCGA cohort and if they were listed in the GSE83300 dataset.
Construction of the prognostic IFN-β-associated gene signature
Univariate Cox regression analysis and lasso-penalized Cox regression analysis were used to identify the prognosis-related IFN-β-associated genes and to construct the prognostic gene signature. P<0.05 in the univariate Cox regression analysis was considered statistically significant. The prognostic gene signature was shown as risk score=(coefficient mRNA1×expression of mRNA1)+(coefficient mRNA2×expression of mRNA2)+…+(coefficient mRNAn×expression mRNAn). R package “survival” and “survminer” were used to explore the optimal cut-off of the risk score and to draw the Kaplan-Meier survival curve. In particular, the “surv_cutpoint” function of the “survminer” R package was used to determine the optimal cut-off value to divide patients into the high- and low-risk groups. R package “survivalROC” was used to investigate the time-dependent prognostic value of the gene signature. A 2-sided log-rank P<0.05 was considered significant for the survival analysis.
IFN-β-associated lncRNA screening
The profiles of the lncRNAs and IFN-β-associated genes were obtained from TCGA RNAseq dataset. Pearson correlation was applied to calculate the correlation between the lncRNAs and differential genes. An lncRNA with a correlation coefficient |R2|>0.3 and P<0.05 was considered to be an IFN-β-associated lncRNA.
Construction of the prognostic IFN-β-associated lncRNA signature
Construction of the prognostic IFN-β-associated lncRNA signature was performed as previously described.
GSEA
GSEA was applied to investigate potential mechanisms underlying the influence of differential gene expression on GBM prognosis. GSEA was also applied to detect whether a priori-defined set of genes showed statistically significant differential expression between the high and low-risk groups. Gene sets with a standard P<0.05 were considered to be significantly enriched.
Immunohistochemical staining
The paraffin-embedded glioma tissues were cut into thin slices and then placed on glass slides for the immunohistochemical experiments. The specimens were incubated with rabbit anti-BCL2L2, anti-PRDX1, anti-XRCC5, and SEC61B antibody (1:200 dilutions; Abcam, Cambridge, MA, USA) at 4 °C overnight, followed by 1-h incubation of biocatalyst secondary antibody (1:200 dilutions, Santa Cruz Biotechnology, Santa Cruz, CA, USA) at room temperature. The avidin-biotin complex method was used to determine the target protein’s location and relative expression to visualize the bound antibodies.
Statistical analysis
R 4.0.2 (www.r-project.org/) and SPSS.22 (www.ibm.com/software/it/analytics/spss/) were used to compute statistical analyses. The association between the IFN-β-associated genes and clinicopathologic features was tested using the chi-square test. Comparison of two independent groups was made by two-tailed Students t test. A one-way analysis of variance (ANOVA) was used to determine differences among groups. Statistical significance was set at *P<0.05, **P<0.01, ***P<0.001. P<0.05 was considered to indicate statistical significance.
Results
Construction and validation of the prognostic IFN-β-associated gene signature
In total, 596 GBM patients and 121 IFN-β-associated genes (70 upregulated and 51 downregulated) were included in TCGA GBM to construct the prognostic model. Differential gene expression analysis identified 14 downregulated and 53 upregulated IFN-β-associated genes, respectively (Table S1). First, the heat map shows the differential genes and analyzed these significant genes further (Figure 1A). Univariate Cox regression model found 5 survival-related genes (Table S2). Lasso-penalized Cox analysis identified 5 genes to construct the prognostic model (Table S3). Using the methodology previously described, the result is validated on the GEO datasets (GSE83300) (Table S4). Patients were divided into high- and low-risk groups depending on their risk score. GBM patients with high-risk scores had poor prognosis (Figure 1B,C). The increased expression of the 4 different signature genes (PRDX1, SEC61B, XRCC5, and Troxerutin (TXN) and reduced expression of the 1 signature gene (BCL2L2) was observed as the risk value increased (Figure 1D,E). Taking all of our results together, 4 genes were found to be correlated with unfavorable clinical outcomes.
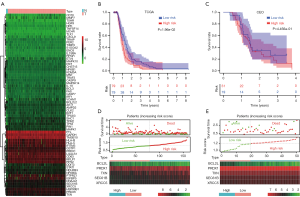
Prognostic significance of the 4 signature gene expression in GBM
To further validate the expression of the prognostic genes constructing the gene signature, Kaplan-Meier survival analysis was used. The findings indicated that the high expression of SEC61B and XRCC5 was associated with poor prognosis in the GEO dataset (Figure 2A,B). Moreover, the high expression of SEC61B, XRCC5, and PRDX1 was associated with poor prognosis, and the low expression of BCL2L2 was associated with poor prognosis in TCGA dataset (Figure 2C,D,E,F). To further verify whether the expression of SEC61B, XRCC5, BCL2L2, and PRDX1 was associated with prognosis in GBM, the Gene Expression Profiling Interactive Analysis (GEPIA) database (https://gepia.cancer-pku.cn/) was used. SEC61B, XRCC5, and PRDX1 had significantly high expression in tumor samples compared with normal samples, and BCL2L2 had significantly low expression in tumor samples compared with normal samples (Figure 2G). GBM samples (n=50) and normal brain tissues (n=50) were then used for real-time polymerase chain reaction (PCR) experiments. The results were consistent with the GEPIA database (Figure 2H). Taken together, the 4 signature gene expression is considered to be of clinical significance in GBM.
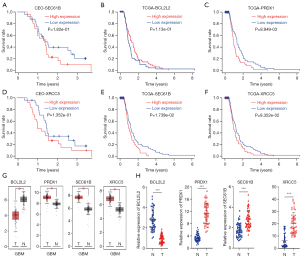
Validation of the 4 signature genes in the CGGA database
To further validate these results, we used the CGGA. Kaplan-Meier survival analysis of the CGGA dataset showed that the high expression of SEC61B, XRCC5, and PRDX1, and the low expression of BCL2L2 indicated poor patient prognosis (Figure 3A,B,C,D). The expression level of BCL2L2 significantly decreased with higher-grade gliomas (Figure 3E). Moreover, the expression of SEC61B, XRCC5, and PRDX1 significantly increased with higher-grade gliomas (Figure 3F,G,H). To further validate these results, we performed immunohistochemical experiments. The immunohistochemical results obtained in the present study were consistent with the results of the CGGA database (Figure 3I). The expression of the 4 signature genes was considered to be of clinical significance in GBM.
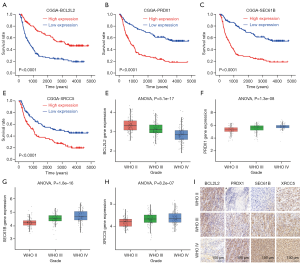
GSEA analysis of the 4 signature genes
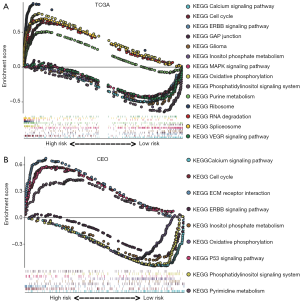
To further clarify the impact of the 4 signature genes on GBM, gene ontology and pathway enrichment analyses were performed using GSEA. The results revealed that these genes are mainly enriched in 14 pathways based on TCGA GBM database, including the calcium signaling pathway, cell cycle, epidermal growth factor receptor family (ERBB) signaling pathway, glyceraldehyde-3-phosphate dehydrogenase (GAP) junction, glioma, inositol phosphate metabolism, mitogen-activated protein kinase (MAPK) signaling pathway, oxidative phosphorylation, phosphatidylinositol signaling system, purine metabolism, ribosome, RNA degradation, spliceosome, and vascular endothelial growth factor (VEGR) signaling pathway (Figure 4A). Moreover, in the GEO dataset, these genes were mainly enriched in 9 pathways as follows: the calcium signaling pathway, cell cycle, extracellular matrix receptor interaction, ERBB signaling pathway, inositol phosphate metabolism, oxidative phosphorylation, P53 signaling pathway, phosphatidylinositol signaling system, and pyrimidine metabolism (Figure 4B). These genes may be involved in the proliferation of GBM.
Prognostic impact of IFN-β-associated lncRNA signature for GBM
Considering the critical role of lncRNAs in GBM, the identification of important lncRNAs in cancer and developing lncRNA-based therapeutic strategies are important. Pearson correlation was applied to calculate the correlation between lncRNAs and IFN-β-associated genes. An lncRNA with a correlation coefficient |R2|>0.3 and P<0.05 was considered to be an IFN-β-associated lncRNA. Univariate and multivariate Cox regressions were used for the survival analysis, and indicated that AC093278.2, AC004067.1, LINC01116, and AC017104.1 were independent prognostic factors for the overall survival of GBM patients (Figure 5A,B,C). Moreover, the high expression of the AC004067.1, AC017104.1, and LINC01116 is associated with poor prognosis, and the low expression of AC093278.2 is associated with poor prognosis in TCGA dataset (Figure 5D,E,F,G). GBM samples (n=50) and normal brain tissues (n=50) were then used for real-time PCR experiments to validate the expression of the IFN-β-associated lncRNAs in GBM. RT-PCR showed that, compared with the normal brain tissues, AC004067.1, AC017104.1, and LINC01116 were highly expressed and AC093278.2 and a low expression in GBM (Figure 5H). Therefore, IFN-β-associated lncRNAs have a high diagnostic value for GBM.
Discussion
With the development of high-throughput sequencing technology, the understanding of cancer is becoming clearer. As the scope of analyzed genes and diseases expands, bioinformatics analysis is becoming increasingly important. In the present study, we analyzed the biological functions of a prognostic IFN-β-associated gene signature using bioinformatics analysis.
Univariate Cox regression model found 5 survival-related genes. Lasso-penalized Cox analysis identified 5 genes to construct the prognostic model. Using the methodology previously described, the result is validated on the GEO datasets (GSE83300). We found that 4 IFN-β-associated genes (PRDX1, SEC61B, XRCC5, and BCL2L2) signature was a suitable marker of seizure prognosis in patients with GBM. Using the CGGA data, we found that 4 IFN-β-associated genes are independent biomarkers of prognosis and play important roles in many biological processes. For example, PRDX1 is a member of the peroxiredoxin family of antioxidant enzymes, which reduce hydrogen peroxide and alkyl hydroperoxides (20). PRDX1 forms a heterodimer with p38α MAPK14, stabilizing phosphate-p38α in glioma cells (21), and epigenetic silencing of PRDX1 is frequent in 1p/19q-deleted oligodendroglial tumors and likely contributes to radiosensitivity and chemosensitivity of these tumors (22). XRCC5 is the 80-kD subunit of the Ku heterodimer protein, which is also known as ATP-dependent DNA helicase II or DNA repair protein XRCC5. The polymorphisms of XRCC5 play an important role in astrocytoma prognosis in the Chinese Han population, which could be used in the determination of astrocytoma prognosis in clinical research (23). Elevated XRCC5 expression level can promote temozolomide resistance and predict poor prognosis in glioblastoma (24). BCL2L2 is a member of the Bcl-2 protein family. The proteins of this family form heterodimers or homodimers and act as anti- and pro-apoptotic regulators. The expression of (24) in various cancer cell types. Interestingly, BCL2L2 mRNA is highly expressed in the mesenchymal type of GBM (25). Through the wide variety of studies published to date, no clear consensus for the BCL2L2 is correlated with radiotherapy and chemotherapy in GBM. SEC61B is the central component of the protein translocation apparatus of the endoplasmic reticulum membrane (26). However, to the best of our knowledge, the expression pattern and function of SEC61B in GBM have not been previously reported; the role of the BCL2L2, XRCC5, SEC61B in glioma radiotherapy still remains unclear. Therefore, further study is warranted.
Considering the critical role of lncRNAs in GBM, the identification of important lncRNAs in cancer and developing lncRNA-based therapeutic strategies will be important in the future. Univariate and multivariate Cox Pearson correlation was applied to calculate the correlation between the lncRNAs and IFN-β-associated genes. regressions were used for the survival analysis, and indicated that AC093278.2, AC004067.1, LINC01116, and AC017104.1 were independent prognostic factors for the overall survival of GBM patients. LncRNA genes play important roles in many biological processes. For example, LINC01116 promotes tumor proliferation, migration, and invasion in glioma cell (27,28). However, the role of AC093278.2, AC004067.1, and AC017104.1 in GBM has not been reported, and it is important that it is elucidated in future studies. All in all, a novel IFN-β-associated gene signature to predict the overall survival of GBM patients, which may help in clinical decision making for individual treatment.
Conclusions
In the present study, we explored and analyzed differentially expressed IFN-β-associated genes by systematic bioinformatics analysis and established a novel IFN-β-associated gene signature to predict the overall survival of GBM patients, which may help in clinical decision making for individual treatment.
Acknowledgments
We are very grateful to all of the participants in the present research and the highly valuable comments of Zhou Jian. We also thank the reviewers for their valuable advice.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-1986
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-1986
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-1986). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of the Jiangxi Cancer Hospital of Nanchang University. Informed consent was obtained from the patients or guardians.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Alexander BM, Ba S, Berger MS, et al. Adaptive Global Innovative Learning Environment for Glioblastoma: GBM AGILE. Clin Cancer Res 2018;24:737-43. [Crossref] [PubMed]
- Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 2007;114:97-109. [Crossref] [PubMed]
- Majc B, Novak M, Jerala NK, et al. Immunotherapy of Glioblastoma: Current Strategies and Challenges in Tumor Model Development. Cells 2021;10:265. [Crossref] [PubMed]
- Oronsky B, Reid TR, Oronsky A, et al. A Review of Newly Diagnosed Glioblastoma. Front Oncol 2021;10:574012 [Crossref] [PubMed]
- Ho AL, Koch MJ, Tanaka S, et al. Impact of histopathological transformation and overall survival in patients with progressive anaplastic glioma. J Clin Neurosci 2016;31:99-105. [Crossref] [PubMed]
- Weller M, van den Bent M, Hopkins K, et al. EANO guideline for the diagnosis and treatment of anaplastic gliomas and glioblastoma. Lancet Oncol 2014;15:e395-403. [Crossref] [PubMed]
- Uddin MS, Mamun AA, Alghamdi BS, et al. Epigenetics of glioblastoma multiforme: From molecular mechanisms to therapeutic approaches. Semin Cancer Biol 2020:S1044-579X(20)30275-3.
- Sulman EP, Ismaila N, Armstrong TS, et al. Radiation Therapy for Glioblastoma: American Society of Clinical Oncology Clinical Practice Guideline Endorsement of the American Society for Radiation Oncology Guideline. J Clin Oncol 2017;35:361-9. [Crossref] [PubMed]
- Azam Z, To ST, Tannous BA. Mesenchymal Transformation: The Rosetta Stone of Glioblastoma Pathogenesis and Therapy Resistance. Adv Sci (Weinh) 2020;7:2002015 [Crossref] [PubMed]
- Peng Y, Pei H. DNA alkylation lesion repair: outcomes and implications in cancer chemotherapy. J Zhejiang Univ Sci B 2021;22:47-62. [Crossref] [PubMed]
- Ochiai Y, Sumi K, Sano E, et al. Antitumor effects of ribavirin in combination with TMZ and IFN-beta in malignant glioma cells. Oncol Lett 2020;20:178. [Crossref] [PubMed]
- GuhaSarkar D, Su Q, Gao G, et al. Systemic AAV9-IFNbeta gene delivery treats highly invasive glioblastoma. Neuro Oncol 2016;18:1508-18. [PubMed]
- Hong YK, Chung DS, Joe YA, et al. Efficient inhibition of in vivo human malignant glioma growth and angiogenesis by interferon-beta treatment at early stage of tumor development. Clin Cancer Res 2000;6:3354-60. [PubMed]
- Natsume A, Aoki K, Ohka F, et al. Genetic analysis in patients with newly diagnosed glioblastomas treated with interferon-beta plus temozolomide in comparison with temozolomide alone. J Neurooncol 2020;148:17-27. [Crossref] [PubMed]
- Makita K, Hara H, Sano E, et al. Interferon-beta sensitizes human malignant melanoma cells to temozolomide-induced apoptosis and autophagy. Int J Oncol 2019;54:1864-74. [PubMed]
- Happold C, Roth P, Silginer M, et al. Interferon-beta induces loss of spherogenicity and overcomes therapy resistance of glioblastoma stem cells. Mol Cancer Ther 2014;13:948-61. [Crossref] [PubMed]
- Statello L, Guo CJ, Chen LL, et al. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 2021;22:96-118. [Crossref] [PubMed]
- Liu B, Zhou J, Wang C, et al. LncRNA SOX2OT promotes temozolomide resistance by elevating SOX2 expression via ALKBH5-mediated epigenetic regulation in glioblastoma. Cell Death Dis 2020;11:384. [Crossref] [PubMed]
- Xu N, Liu B, Lian C, et al. Long noncoding RNA AC003092.1 promotes temozolomide chemosensitivity through miR-195/TFPI-2 signaling modulation in glioblastoma. Cell Death Dis 2018;9:1139. [Crossref] [PubMed]
- Kisucka J, Chauhan AK, Patten IS, et al. Peroxiredoxin1 prevents excessive endothelial activation and early atherosclerosis. Circ Res 2008;103:598-605. [Crossref] [PubMed]
- Wirthschaft P, Bode J, Simon AEM, et al. A PRDX1-p38alpha heterodimer amplifies MET-driven invasion of IDH-wildtype and IDH-mutant gliomas. Int J Cancer 2018;143:1176-87. [Crossref] [PubMed]
- Dittmann LM, Danner A, Gronych J, et al. Downregulation of PRDX1 by promoter hypermethylation is frequent in 1p/19q-deleted oligodendroglial tumours and increases radio- and chemosensitivity of Hs683 glioma cells in vitro. Oncogene 2012;31:3409-18. [Crossref] [PubMed]
- He X, Zhu X, Li L, et al. The relationship between polymorphisms of XRCC5 genes with astrocytoma prognosis in the Han Chinese population. Oncotarget 2016;7:85283-90. [Crossref] [PubMed]
- Lee IN, Yang JT, Huang C, et al. Elevated XRCC5 expression level can promote temozolomide resistance and predict poor prognosis in glioblastoma. Oncol Lett 2021;21:443. [Crossref] [PubMed]
- Chung HJ, Choi YE, Kim ES, et al. miR-29b attenuates tumorigenicity and stemness maintenance in human glioblastoma multiforme by directly targeting BCL2L2. Oncotarget 2015;6:18429-44. [Crossref] [PubMed]
- Zhu Y, Zhang G, Lin S, et al. Sec61beta facilitates the maintenance of endoplasmic reticulum homeostasis by associating microtubules. Protein Cell 2018;9:616-28. [Crossref] [PubMed]
- Wang T, Cao L, Dong X, et al. LINC01116 promotes tumor proliferation and neutrophil recruitment via DDX5-mediated regulation of IL-1beta in glioma cell. Cell Death Dis 2020;11:302. [Crossref] [PubMed]
- Zhang N, Shuai K, Cheng J, et al. LncRNA linc01116 prometes glioma cell migration and invasion by modulation of radixin targeted by miR-31. Int J Clin Exp Pathol 2019;12:1078-86. [PubMed]
(English Language Editor: R. Scott)


