
Characterization of comprehensive dynamic epigenetic changes during human primary Sjögren’s syndrome progression
Introduction
Primary Sjögren’s syndrome (pSS) is a systemic autoimmune disease characterized by a reduction in the function of exocrine glands, mainly salivary and lacrimal glands, caused by chronic lymphocytic infiltration, elevated proinflammatory cytokine levels, and circulating autoantibodies (1). Based on the classification criteria of the American-European Consensus Group, pSS has a prevalence of approximately 0.4% in the general population and is far more prevalent in females than in males (2). The main clinical manifestations of pSS are dry mouth and dry eyes. Xerostomia is the foremost oral manifestation, presenting in 94% of patients with SS (3). pSS progression can also lead to more problems involving the oral cavity, including atrophy of the tongue papillae, abnormal taste sensation, oral pain, pararthria, dysphagia, oral mucosal ulcers, periodontal disease, oral fungal infections, and parotid enlargement. Some reports have assessed the alterations in voice, speech, swallowing, smell, and taste as well as their effects on the quality of life (4). Moreover, approximately 30–50% of pSS patients have a significantly elevated risk of extra-glandular manifestations including non-Hodgkin lymphoma of the B-cell series, such as mucosa-associated lymphoid tissue (MALT) lymphoma (5,6). Clinically, pSS is characterized by elevated rheumatoid factor (RF) levels, erythrocyte sedimentation rate (ESR), and the production of both organ-specific and nonspecific autoantibodies, including anti-SSA, anti-SSB, anti-nuclear antibody (ANA), and anti-centromere antibodies (ACA) (7). Although both the etiology and the pathogenesis of pSS remain unclear, genetic predispositions, environmental factors, and epigenetic mechanisms can contribute to disease progression.
Epigenetics involves inheritable and reversible processes that govern gene expression without altering the DNA sequence. Histones with N-terminal lysine, arginine, and serine residues that are regulated by multiple chemical modifications have been well-documented as indicators of DNA methylation and chromatin accessibility that affect gene expression. Previous studies have indicated that pSS susceptibility variants are specifically associated with mono-methylated histone 3 lysine 4 (H3K4me1), acetylated histone 3 lysine 27 (H3K27ac), and tri-methylated histone 3 lysine 36 (H3K36me3) for promoters and enhancers in salivary gland cells and immunocytes (8,9). However, dynamic profiles of other covalent histone modifications are unknown in pSS occurrence and progression.
In this study, we harvested human salivary gland tissues from pSS patients with MALT lymphoma and conducted RNA sequencing (RNA-seq) and chromatin immunoprecipitation sequencing (ChIP-seq) for tri-methylated histone 3 lysine 4, 9, 27, 36 and 79 (H3K4/9/27/36/79me3) to investigate the dynamic changes and coordinated interplays of these complex epigenetic codes, thereby elucidating their influence on gene expression during the progression from pSS to MALT lymphoma. Our high-throughput data were valuable in highlighting the importance of epigenetic alterations in the pathogenesis of pSS, as well as identifying potential targets for therapeutic intervention of pSS. This article has been presented in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/atm-21-1754).
Methods
Clinical specimen harvesting and handling
Two female patients admitted to the Department of Oral and Maxillofacial Surgery at the Ninth People’s Hospital, School of Medicine, Shanghai Jiao Tong University, between 2012 and 2018 were enrolled in this study. They were diagnosed with pSS based on the classification criteria of American-European Consensus Group (2002 version) (10). Both patients had their labial gland tissues harvested to examine lymphocyte infiltration at the first diagnosis of pSS; their parotid glands with and without lymphoma lesions were excised followed by the second diagnosis of local MALT lymphoma. The tissues to be used in subsequent experiments were stored at −80 °C immediately after harvesting. This study was approved by the Committee of Ethics, Faculty of Medicine, Shanghai Jiao Tong University (SH9H-2019-T159-2). Informed consent was obtained from all participants. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
RNA-seq data processing
Total RNA was extracted and the quality of all samples was evaluated using an Agilent bioanalyzer 2100 (Agilent, Santa Clara, CA, USA). The samples had an average RNA integrity number (RIN) of 9.4 with a minimum of 7.5. From each sample, 5 μg of RNA was used for library preparation using the NEBNext Ultra Directional RNA Library Prep Kit by Illumina (NEB, Ipswich, MA, USA), following the manufacturer’s instructions. Libraries were pooled and PE150 was sequenced using the Illumina HiSeq 2500 platform. Raw read quality was evaluated using FastQC. Adapters and low-quality bases with a quality score of lower than 15 were trimmed from the raw RNA-seq reads. After trimming, reads shorter than 30 bp were discarded. The clean reads were aligned with the human reference genome hg38 using STAR (2.3.0) and assembled and quantified using Hisat2 (0.5.4p5). The differentially expressed genes (DEGs) were identified using the DESeq2 package in R. In order to be considered a DEG, there had to be a two-fold change in expression levels between samples adjusted using the Benjamini & Hochberg (BH) method. Transcription levels were then converted to logarithms (log2) of the normalized counts. Gene annotation was described using the Ensembl genome browser database (http://www.ensembl.org/index.html). The ClusterProfiler package in R and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were used to annotate the DEGs with gene ontology (GO) terms. P values below 0.05 were considered statistically significant.
ChIP-seq data processing
The sample containing 5×106 cells was ground in liquid nitrogen, dissolved in lysis buffer [20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton, 2.5 mM sodium pyrophosphate, 1 mM beta-glycerophosphate, 1 mM Na3VO4, 1 µg/mL leupeptin, 1 mM PMSF, and 1 mM protease inhibitor cocktail (Solarbio, Beijing, China)] and placed on ice for 30 min. After the genomic DNA was broken into 200–500 bp fragments by sonication, 10% of each whole lysate was saved. The rest of the lysate was divided into five parts, dissolved into 1 mL of lysis buffer, and incubated overnight with 1 μg IP-grade antibodies against H3K4me3 (Cat. No. 9751, Cell Signaling Technology, Beverly, MA, USA), H3K9me3 (Cat. No. 13969, Cell Signaling Technology), H3K27me3 (Cat. No. 9733, Cell Signaling Technology), H3K36me3 (Cat. No. 4909, Cell Signaling Technology), or H3K79me3 (Cat. No. 74073, Cell Signaling Technology). This was followed by a protein-A beads pull down at 4 °C for 2 h. The DNA adhered to the beads as well as the input DNA were purified using phenol chloroform, and the concentration of purified DNA was determined using the Nano Drop 2000 (Thermo Fisher Scientific, Waltham, MA, USA). The DNA was repaired up to the 3' dA overhang and then ligated with adapters. The libraries were assembled after eliminating the unligated adapters and an appropriate size was selected for SE75 sequencing using the Illumina Hiseq2500 platform. After trimming adaptors from the raw ChIP-seq reads and filtering out low-quality reads, the remaining reads were mapped to the hg38 genome using the Bowtie 2 (v2.2.6) algorithm. Peaks were called using MACS2 (2.1.1) with a P value cutoff of 0.01, the different binding domains were analyzed based on an FDR value of less than 0.05 and annotated using DiffBind. The peaks on certain genomic loci were visualized using the Integrative Genomics Viewer (IGV). GO and KEGG analyses were performed as described in the previous section.
Data availability
The raw data that support the findings of this study have been deposited in the ArrayExpress database (https://www.ebi.ac.uk/arrayexpress/) and assigned with an accession number E-MTAB-10339/10340. Any other data are available from the corresponding authors upon request.
Results
Transcriptome profiling in the salivary gland cells of pSS patients
The two female patients we focused on in this study were diagnosed with pSS and mainly presented with oral symptoms without any other complications such as cryoglobulinemia, hypergammaglobulinemia, or allergic purpura. The age at diagnosis of pSS was 35 years for patient A and 54 years for patient B. The focus score for patient A was 1 and that for patient B was 2. There was no Epstein Barr virus infection in either patient. The oral dryness VAS score for patient A was 5 and that for patient B was 6. The age at diagnosis of MALT was 37 years for patient A and 57 years for patient B. Both patients A and B had an oral dryness VAS score of 6. At the diagnosis of pSS and MALT, both anti-SSA and anti-SSB were positive; RF levels and ESR were abnormally elevated in both patients. The detailed results are presented in Table 1.
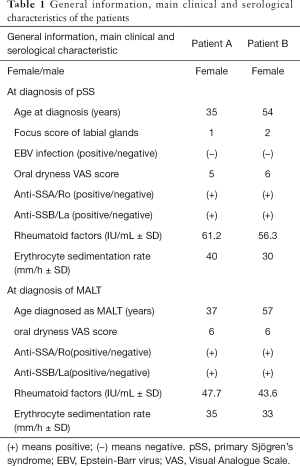
Full table
Two repeated sequencing libraries were established from the labial gland tissue samples from both pSS patients with (LG_pSS) or without (LG_NC) lymphocyte infiltration. Two repeated sequencing libraries were established from the parotid glands with (PG_MALT) and without (PG_NC) MALT lymphoma tissues for these two patients who eventually developed MALT lymphoma. In order to broadly assess transcriptome alterations and changes in epigenetic regulation of gene expression during pSS progression, RNA-seq and ChIP-seq were conducted and the data were analyzed (Figure 1A). The alignment of data and the correlations with duplications in each group are summarized in Table S1, which indicated that only biological materials with the best quality were used in this study.
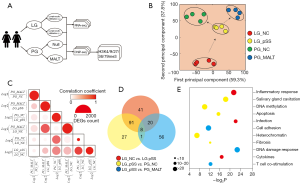
Principal component analysis (PCA) of the RNA-seq data indicated that four distinct clusters of samples could be separated by the first and second principal components (Figure 1B). As recently described in extranodal marginal zone lymphoma of MALT type, the vast proportion of cells were still defined as gland cells (11). Therefore, RNA-seq data could truly reflect the expression profile of gland cells in PG_MALT. Apart from LG_NC, the other three clusters were more relevant to each other, indicating more similar transcriptome patterns between the gland cells from pSS and MALT compared to those from the negative control. The expression profiles also suggested that PG_NC and PG_MALT seemingly had a completely different direction of pSS progression from that of LG_pSS, differing based on the first principal component. PG_NC harvested from the second diagnosis of the same pSS patients were behind LG_pSS in terms of time course. Therefore, it was reasonable to conclude that PG_NC and PG_MALT might have resulted from pSS progression.
Moreover, given the differences between the treated samples and the control, we also performed a comparative analysis among four clusters of samples that included 1,236 DEGs between LG_pSS and LG_NC, 233 DEGs between PG_NC and LG_NC, 1,557 DEGs between PG_MALT and LG_NC, 1,427 DEGs between PG_NC and LG_pSS, 1,516 DEGs between PG_MALT and LG_pSS, and 1,244 DEGs between PG_MALT and PG_NC. We then obtained correlation coefficients by comparing the aforementioned DEGs (Figure 1C). We noticed that there were two routes: the first from LG_NC to LG_pSS to PG_NC (Route 1); the second from LG_NC to LG_pSS to PG_MALT (Route 2).Both routes demonstrated a relatively tight correlation among each other, whereas the global transcriptome changes across these two developmental routes had a lesser correlation. Although DEGs between PG_NC vs. PG_MALT were large in number, their correlation with DEGs from other comparisons was extremely low.
Finally, GO analysis of intersected DEGs (Figure 1D) revealed that GO terms of “inflammatory response”, “infection”, and “cytokines” were overlapped across Routes 1 and 2. DNA methylation, heterochromatin, cell apoptosis, cavitation, and fibrosis particularly occurred on Route 1, whereas DNA damage response, cell adhesion, and T cell co-stimulation only occurred on Route 2 (Figure 1E).
The above results indicated two putative outcomes of pSS progression involving a large number of different genes and functions.
Dynamic profiles of covalent histone modifications during pSS progression
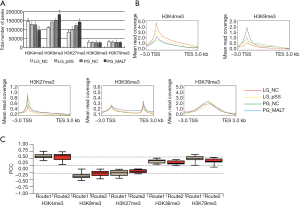
Next, we focused on the tri-methylation of histone 3 at specific lysine residues to explore the mechanism underlying transcriptional regulation during pSS progression. Overall trends for H3K36/79me3 were not evident; the genome-wide peaks for H3K4me3 were significantly downregulated, whereas those for H3K9/27me3 were upregulated during pSS progression (Figure 2A). Furthermore, metagene profiles displayed on tiled regions revealed that the H3K4/9/27me3 enrichments were majorly on the promoters, and the H3K36me3 also appeared at the 3' untranslated regions (3' UTRs) next to the promoters, whereas H3K79me3 only existed in the gene body (Figure 2B). We specifically focused on the DEGs from Routes 1 and 2 in order to investigate the relevance of epigenetic regulation. The Pearson correlation between changes in gene expression and the different histone patterns was calculated. There was a strong positive correlation between H3K4me3 and gene expression in Routes 1 and 2, as well as a strong negative correlation between H3K9me3 and gene expression in Route 1 (r≥0.3 or ≤−0.3), whereas the other correlations were relatively weak (−0.3<r<0.3) (Figure 2C). All the analyzed correlations were statistically significant (P<0.05), suggesting that these epigenetic modifications might contribute to modulating gene expression during pSS progression.
Causality and priority of histone modifications in pSS pathogenesis
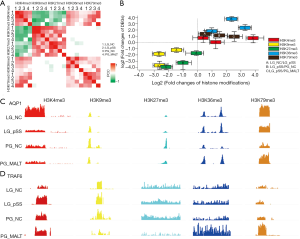
We further compared the correlations among the histone modifications for all DEGs (Figure 3A). Interestingly, our observations indicated potential connections among histone modifications. First, H3K4me3 was roughly associated with H3K9/27/36/79me3 for all DEGs, which implied that H3K4m3 was the most extensively used epigenetic regulation mechanism for gene expression on the genomic scale. Second, the enrichment of H3K9/27me3 shared one group of DEGs, whereas H3K36/79me3 concurrently appeared on another population of DEGs; these two groups of DEGs had no tight relevance with each other. For the comparison between PG_NC and PG_MALT, all the histone modifications displayed poor relevance. Moreover, we analyzed the temporal effects of these five histone modifications on the change in expression for all DEGs in Route 1 and Route 2 of pSS progression (Figure 3B).H3K4me3 was the first histone modification from LG_NC to LG_pSS, followed by H3K9me3, H3K36me3, H3K79me3, and H3K27me3. Unexpectedly, H3K27me3 and H3K36me3 were likely to contribute to the most powerful force and regulate gene expression at the stages from LG_pSS to PG_NC. Likewise, H3K9me3 and H3K27me3 majorly impacted the expression of DEGs at the stages from LG_pSS to PG_MALT. The gene browser views of AQP1 [log2(fold change) =−2.53, P<0.001] (Route 1) and TRAF6 [log2(fold change) =1.82, P<0.001] (Route 2), were exemplified to verify this novel regulatory model (Figure 3C,D). These results demonstrate that H3K9/27/36/79me3 could control the expression of a specific group of DEGs based on the genome-wide layout of H3K4me3 in pSS.
Discussion
A previous retrospective study has suggested that MALT lymphoma is a significant yet minor portion of pSS in salivary gland lymphoma (12), indicating that MALT lymphoma is one possible rather than the inevitable ending of pSS. Our study determines that even in one individual, the expression profiles in the salivary glands suffering from pSS exhibit heterogenous outcomes as time goes on (Figure 1B). We presume that it may depend on the surrounding status of chronic inflammation caused by the invasion of lymphocytes. However, our analysis suggests that the transcriptome landscapes and involved biological functions of PG_NC and PG_MALT were less associated with each other (Figure 1C,E). The secretory function of salivary glands, which can be used to reliably evaluate the severity and classification of pSS (13), supports our GO enrichment analysis, wherein salivary glands atrophy and cavitation usually appear in PG_NC but not in the early stage of LG_pSS, as suggested by reduced expression of CCND2 (14), SFRP1 (15), and IGF-1 (16). While the secretory function may not be appropriate to be used to evaluate the glands with MALT lymphoma, parotid gland swelling accompanied with recurrent infection and persistent pain is highly associated with MALT, displaying irregular expression of IL-17 (17) and BCL-2 (18). Consistently, we observed that H3K9me3 and H3K27me3 of Bcl-2 significantly decreased in PG_MALT compared to those in PG_NC and LG_pSS, indicating that histone alterations indeed influence gene expression in pSS progression. Moreover, both cell adhesion and DNA damage response imply that these gland cells with MALT lymphoma escape from programmed cell death and are potentially inclined to undergo a malignant change. An important finding of this study is that the different outcomes of pSS progression can be distinguished based on the distinct global transcription patterns.
Of various histone codes, we mainly focused on the dynamic changes and interplays for tri-methylation of histone 3 at lysine residues 4, 9, 27, 36, and 79 during pSS development. Among active genes, general observations include H3K4me3 at the promoters (19), H3K36me3 at the promoters and the 3' UTR (20), as well as H3K79me3 enriched in the gene body (21). H3K9me3 and H3K27me3 are linked to transcriptional repression (22). The overall trends of H3K4me3 decrease and H3K9/27me3 increase have been documented characterizations of heterochromosome and gene silencing in salivary gland cells with pSS progression irrespective of the direction in which they go (Figure 2A,B). H3K4/9/27me3 can be used as important histone benchmarks to determine the severity of pSS. Although genome-wide alterations have no difference, significant correlations with gene transcription indicate that the changes in H3K36/79me3 become balanced with the concurrent up and downregulation of target genes (Figure 2C). Histone lysine methylation is coordinated by the histone methyltransferase (HMT) and demethylase (KDM) families that can specifically recognize methylated residues. These enzymes may be crucial in some important life processes or play a bridging role in the regulation of chromatin function. Based on the limited data, we were unable to determine the activities of these enzymes in pSS; however, we assume that small molecular inhibitors of HTM or HDM are likely to relieve the severity of pSS, which needs to be explored in future studies.
The interplay among different histone modifications in our findings suggests that H3K4me3 correlates with other four histone modifications and is a fundamental epigenetic modification for transcriptional regulation on the genomic scale (Figure 3A), which is consistent with the previous studies on the crosstalk between H3K4me3 and H3K27me3 (23),H3K36me3 (24) as well as H3K79me3 (25). Moreover, the change in H3K4me3 is among the earliest events in the pathological process from LG_NC to LG_pSS; however, it is not the primary contributor to gene transcription changes (Figure 3B). The most likely explanation is that H3K4me3 alteration might create a unique chromosomal environment of mutual exclusivity for H3K9/27me3 or a synergetic effect on H3K36/79me3 to facilitate the recruitment of more epigenetic regulators and transcription factors for the target gene. Therefore, a variation in H3K4me3 can be considered as an advanced diagnostic indicator for early intervention at the initial stage of pSS in clinical trials. H3K27me3 and H3K36me3 seemingly play crucial roles in regulating gene activation or silencing to result in the outcomes of pSS progression (Figure 3B). Finally, selective changes in H3K9/27/36/79me3 enrichments on target genes in PG_NC or PG_MALT determine the final progression of Route 1 or Route 2 (Figure 3C,D). The mechanism underlying this interesting discovery needs to be further investigated by studying the specific functions of multiple epigenetic regulatory enzymes in gland cells.
There are still certain shortcomings in our study such as the limited sample size as well as the less abundant types of pSS. Besides MALT lymphoma, other extraglandular complications of pSS are likely to reflect other routes of pSS progression. Moreover, identification of more epigenetic signatures may help advance our understanding of the molecular mechanisms involved in pSS pathogenesis and progression. The target genes and signaling pathways affected by different epigenetic modifications will be validated in future studies in order to develop therapeutic strategies through epigenetic intervention in pSS.
Conclusions
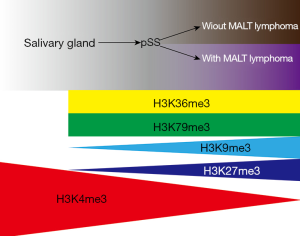
Overall, our study reveals a novel pathological change in pSS from the global transcriptome alterations regulated by a coordinated epigenetic mechanism (Figure 4). Our study advances the understanding of pSS pathogenesis, gives an insight into the putative molecular theory for pSS progression, and provides a potential therapeutic strategy for pSS treatment.
Acknowledgments
We appreciate the linguistic assistance provided by TopEdit (
Funding: This work was supported by National Natural Science Foundation of China (Grant No. 81671004, 81771089, 81800990).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-1754
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-1754
Peer Review File: Available at https://dx.doi.org/10.21037/atm-21-1754
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-1754). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Committee of Ethics, Faculty of Medicine, Shanghai Jiao Tong University (SH9H-2019-T159-2), and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Feltsan T, Stanko P, Mracna J. Sjogren s syndrome in present. Bratisl Lek Listy 2012;113:514-6. [PubMed]
- Westhoff G, Zink A. Epidemiology of primary Sjorgren's syndrome. Z Rheumatol 2010;69:41-9. [Crossref] [PubMed]
- Ruiz Allec LD, Hernandez Lopez X, Arreguin Porras JB, et al. Alterations in voice, speech and swallowing in patients with Sjogren's syndrome. Acta Otorrinolaringol Esp 2011;62:255-64. [Crossref] [PubMed]
- Kamel UF, Maddison P, Whitaker R. Impact of primary Sjogren's syndrome on smell and taste: effect on quality of life. Rheumatology (Oxford) 2009;48:1512-4. [Crossref] [PubMed]
- Stefanski AL, Tomiak C, Pleyer U, et al. The Diagnosis and Treatment of Sjogren's Syndrome. Dtsch Arztebl Int 2017;114:354-61. [Crossref] [PubMed]
- Odani T, Chiorini JA. Targeting primary Sjogren's syndrome. Mod Rheumatol 2019;29:70-86. [Crossref] [PubMed]
- Kyriakidis NC, Kapsogeorgou EK, Tzioufas AG. A comprehensive review of autoantibodies in primary Sjogren's syndrome: clinical phenotypes and regulatory mechanisms. J Autoimmun 2014;51:67-74. [Crossref] [PubMed]
- Konsta OD, Le Dantec C, Charras A, et al. An in silico Approach Reveals Associations between Genetic and Epigenetic Factors within Regulatory Elements in B Cells from Primary Sjogren's Syndrome Patients. Front Immunol 2015;6:437. [Crossref] [PubMed]
- Bordron A, Devauchelle-Pensec V, Le Dantec C, et al. Epigenetics in Primary Sjogren's Syndrome. Adv Exp Med Biol 2020;1253:285-308. [Crossref] [PubMed]
- Vitali C, Bombardieri S, Jonsson R, et al. Classification criteria for Sjogren's syndrome: a revised version of the European criteria proposed by the American-European Consensus Group. Ann Rheum Dis 2002;61:554-8. [Crossref] [PubMed]
- Zhang C, Xia R, Gu T, et al. Clinicopathological aspects of primary mucosa-associated lymphoid tissue lymphoma of the salivary gland: A retrospective single-center analysis of 72 cases. J Oral Pathol Med 2021; Epub ahead of print. [Crossref] [PubMed]
- Travaglino A, Giordano C, Pace M, et al. Sjogren Syndrome in Primary Salivary Gland Lymphoma. Am J Clin Pathol 2020;153:719-24. [Crossref] [PubMed]
- Blokland SLM, Hillen MR, van Vliet-Moret FM, et al. Salivary gland secretome: a novel tool towards molecular stratification of patients with primary Sjogren's syndrome and non-autoimmune sicca. RMD Open 2019;5:e000772 [Crossref] [PubMed]
- Carpenter GH, Cotroneo E. Salivary gland regeneration. Front Oral Biol 2010;14:107-28. [Crossref] [PubMed]
- Lee CH, Hung YJ, Lin CY, et al. Loss of SFRP1 expression is associated with aberrant beta-catenin distribution and tumor progression in mucoepidermoid carcinoma of salivary glands. Ann Surg Oncol 2010;17:2237-46. [Crossref] [PubMed]
- Paszynska E, Dmitrzak-Weglarz M, Slopien A, et al. Salivary and serum insulin-like growth factor (IGF-1) assays in anorexic patients. World J Biol Psychiatry 2016;17:615-21. [PubMed]
- Alunno A, Bistoni O, Caterbi S, et al. Serum interleukin-17 in primary Sjogren's syndrome: association with disease duration and parotid gland swelling. Clin Exp Rheumatol 2015;33:129. [PubMed]
- Hjelmervik TO, Petersen K, Jonassen I, et al. Gene expression profiling of minor salivary glands clearly distinguishes primary Sjogren's syndrome patients from healthy control subjects. Arthritis Rheum 2005;52:1534-44. [Crossref] [PubMed]
- Barrera LO, Li Z, Smith AD, et al. Genome-wide mapping and analysis of active promoters in mouse embryonic stem cells and adult organs. Genome Res 2008;18:46-59. [Crossref] [PubMed]
- Pokholok DK, Harbison CT, Levine S, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 2005;122:517-27. [Crossref] [PubMed]
- Ng HH, Ciccone DN, Morshead KB, et al. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc Natl Acad Sci U S A 2003;100:1820-5. [Crossref] [PubMed]
- Zhang T, Cooper S, Brockdorff N. The interplay of histone modifications - writers that read. EMBO Rep 2015;16:1467-81. [Crossref] [PubMed]
- Douillet D, Sze CC, Ryan C, et al. Uncoupling histone H3K4 trimethylation from developmental gene expression via an equilibrium of COMPASS, Polycomb and DNA methylation. Nat Genet 2020;52:615-25. [Crossref] [PubMed]
- He J, Kallin EM, Tsukada Y, et al. The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b). Nat Struct Mol Biol 2008;15:1169-75. [Crossref] [PubMed]
- Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 2002;418:104-8. [Crossref] [PubMed]


