
Identification and validation of potential novel biomarkers to predict distant metastasis in differentiated thyroid cancer
Introduction
Differentiated thyroid cancer (DTC) is the most common histological subtype of thyroidal malignancies and its incidence has been significantly increasing worldwide in recent years (1,2). DTC has a favorable prognosis, and the rates of 10-year overall survival and disease-specific survival are 89.5–90.8% and 96.6–98.6% (3), respectively. However, 1–9% of DTC patients present distant metastasis (DM) at initial diagnosis (4,5), DM dramatically lowers the survival rate and is the main cause of thyroid cancer-related mortality. Therefore, identifying the molecular profiles and risk factors of DM is pivotal for further in improving the survival rate of the patients.
Of late, the weighted gene co-expression network analysis (WGCNA) algorithm is gaining popularity in constructing the interactive connections between clinical traits and gene modules, WGCNA establishes a scale-free gene co-expression networks, which are an effective in investigating the regulatory mechanisms behind a certain characteristics (6). More importantly, WGCNA is a powerful genetic analysis tool to select new therapeutic targets and candidate biomarkers. Some studies have successfully used WGCNA to investigate hub genes and functional co-expression modules in many diseases, such as obesity (7), bronchopulmonary dysplasia (8), hepatocellular carcinoma (9,10), osteoporosis (11) and sepsis (12). However, the role of the network in patients with DTC and its potential mechanism are yet to be elucidated.
Previous studies on DTC with DM have focused only on the clinicopathological characteristics or gene mutations. Some studies (4,13,14) have demonstrated that lymph node metastasis (LNM) status, tumor size, and extrathyroidal extension (ETE) are the independent risk factors for DM. Other studies (15,16) have employed genomic sequence analysis to elucidate the complex molecular mechanisms of metastatic DTC. TERT promoter, PLEKHS1 promoter, histone methyltransferases, and TP53 have been revealed to be significantly associated with DM, and the RAS mutation has been found to be the predominant driver of DM (17). However, these pathological factors and gene signatures cannot accurately predict the risk of DM.
To fill this knowledge gap, we applied the WGCNA to identify the highly connected hub genes and core modules associated with DM in DTC. Subsequently, we explored the clinicopathological factors by using The Cancer Genome Atlas (TCGA) database to verify the independent risk factors. Finally, we established an integrated nomogram prediction model. To the best of our knowledge, the present study is the first to utilize a nomogram to effectively integrate hub genes with clinical risk factors to predict DM. The tool can provide a new individual risk assessment of the DM in patients with DTC and assist in clinical decision-making. We present the following article in accordance with the TRIPOD reporting checklist (available at https://dx.doi.org/10.21037/atm-21-383).
Methods
Data acquisition and processing
We downloaded the RNA-seq dataset containing 502 thyroid cancer and 58 normal samples from TCGA database (https://portal.gdc.cancer.gov). Furthermore, we downloaded the matched clinical dataset (including gender, age, T stage, histological type, ETE, and DM) from University of California Santa Cruz (UCSC) Xena (http://xena.ucsc.edu/). The differentially expressed genes (DEGs) between the normal tissues and DTC were screened using the “limma” package in R software, The Fragments Per Kilobase Million (FPKM) was used to quantify the expressions of genes. Genes with expression values <0.5 were filtered out, and the remaining 9542 DEGs were selected for further WCGNA analysis. The microarray dataset GSE60542, comprising 45 DTC tissues and 23 normal thyroid tissues, was downloaded from Gene Expression Omnibus (GEO). This dataset is based on the GPL570 (HG-U133_Plus_2) Affymetrix Human Genome U133 Plus 2.0 Array (18), and was used to confirm whether the real hub genes downloaded from TCGA database were reproducible. A flow chart of the present study is depicted in Figure 1. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Gene co-expression network analysis
The R package of WGCNA was utilized to construct a gene co-expression network based on the DEGs. First, a similarity matrix was constructed using Pearson correlation analysis. Second, a soft-thresholding power β was selected to make the similarity matrix achieve an approximate scale-free network topology. Third, the adjacency matrix of the gene expression data was clustered by using topological overlap matrix (TOM) analysis. Finally, the dynamic tree cut algorithm was used to detect co-expressing gene modules (minModuleSize =50). Highly similar modules were merged using 0.25 as the cut-off value.
Identification of modules significantly associated with DM
Module eigengenes (MEs) represents the major principal component of each gene module, and describes the expression patterns of all genes (19). To identify the modules significantly associated with DM, the relationship between MEs and DM was calculated using the Pearson’s correlation test. The modules most significantly related to DM were considered as the key modules for further analysis.
Functional and pathway enrichment analyses
To gain further insights into the functions of the modules related to DM, gene ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed. These analyses were used to annotate the molecular functions, biological processes, cellular components, and abnormal pathways.
Screening the candidate hub genes
Module membership (MM) signifies the relationship between the ME and gene expression. Gene significance (GS) denotes the correlation between the clinical traits and the expression levels of certain genes. An ideal hub gene contains genes of high correlation between MM and GS. In the present study, the hub genes were identified based on the absolute values of GS >0.4 and MM >0.85 and P values <0.05. To verify whether a hub gene is significantly associated with DM, receiver operating characteristic (ROC) curve was applied to validate the diagnostic value. Real hub genes were identified based on the area under the ROC curve (AUC) of >0.75.
Nomogram model development
To identify the clinical risk factors of DM, univariate and multivariate logistic regression analyses were performed to identify the clinical features related to DM. Subsequently, the R package “forestplot” was used to construct the plot and draw the correlation coefficient graph. To improve the risk stratification and assist in the clinical procedures, a nomogram model was developed based on clinicopathological risk factors and gene signatures. The predictive performance of the nomogram was validated using calibrated plots and concordance index (C index). Decision curve analysis (DCA) was employed to assess the clinical values and predictive accuracies. The R packages “survcomp”, “survival”, “stdca.R”, “rms”, and “timeROC” were applied for the analysis.
Quantitative real-time polymerase chain reaction validation
Four human thyroid cancer cell lines (B-CPAP, K1, TPC-1 and KTC-1) and one human normal thyroid epithelial cell line (Nthy-ori3-1) were used in this study. The cells were cultured in a complete medium composed of 10% fetal bovine serum (Gibco) and 90% RPMI1640 (Gibco, Carlsbad, USA), which was supplemented with 100 mg/mL streptomycin and 100 U/mL penicillin (HyClone). The cells were incubated in 5% CO2 at 37 °C and were sub cultured for 10–20 times.
Total RNA was extracted using the AG RNAex Pro RNA extraction kit (AG21101). The RNA was reversely transcribed using PrimeScript™ RT Reagent kit. Reverse transcription was used to compose the first-strand cDNA using 100 ng of total RNA according to the manufacturer’s instructions. Subsequently, quantitative real-time polymerase chain reaction (qRT-PCR) analysis was carried out using the TBGreen Premix Ex TaqTMII (Cat # RR047A-5, TaKaRa, Japan). GAPDH was used as internal control. Primer sequences of the related hub genes are listed in Supplementary Table S1. Relative expression levels were calculated as ratios normalized to GAPDH. The Cq-value for each sample was calculated using the ΔΔCq method, and the results were expressed as 2-ΔΔCq. Real hub genes were identified based on whether the qRT-PCR results exhibited significant differences. The experiments were performed in triplicates.
Statistical analysis
Statistical evaluations were performed using the RStudio software (version 3.5.3). Categorical and continuous variables compared using the Chi-squared test and student’s t-test, respectively. Univariate and multivariate logistic regression analyses were employed to the identify independent factors associated with DM. The P value <0.05 was considered statistically significant.
Results
Weighted co-expression network construction
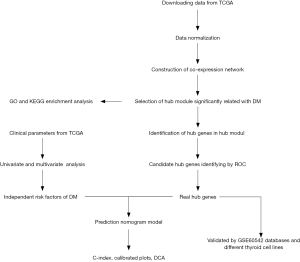
A total of 9,542 differentially expressed mRNAs were used to construct the weighted co-expressed networks. When a soft thresholding power value of 5 was used, the correlation coefficient threshold reached 0.85 and ensured that the topology network to satisfied the scale-free criterion (Figure 2A). The dynamic tree cutting algorithm identified 27 merged co-expression modules, and each one was illustrated using a distinct color (Figure 2B). The number of genes in each module is shown in Table S2. Four MEs, namely greenyellow, lightyellow, midnightblue and red, were significantly positively related DM based on the hierarchical clustering analysis (Figure 2C). Furthermore, the midnightblue module was most significantly positively related to DM. On the other hand, the greenyellow module was negatively with DM, with a correlation of 0.27. Based on the threshold criteria of P<0.05 and |r|>0.4, the midnightblue module was inferred to be the key module associated with DM. Hence, this module was selected for further analysis. The module showed a high positive correlation to DM in patients with DTC (Figure 2D). Next, we demonstrated the TOM among the genes by using the heatmap plot, White color alluded a high overlap, while red color signified a low overlap (Figure 2E). Furthermore, the heatmap plot of the topological overlap revealed the eigengene adjacency of the modules, which was labeled using different colors (Figure 2F).
Functional and pathway enrichment analysis
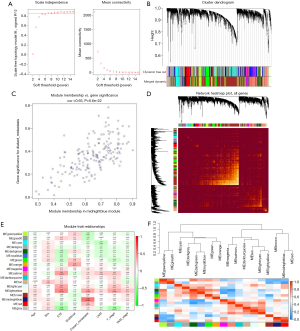
GO enrichment analysis was utilized to explore the biological functions of the genes in the midnightblue module. The top five enriched in components were connective tissue development, endoplasmic reticulum lumen, autophagosome, skeletal system morphogenesis, and transferase activity. KEGG analysis revealed that metabolic pathways,biosynthesis of antibiotics, protein digestion and absorption were the top three pathways in which the midnightblue module genes were involved (Figure 3).
Identification of real hub genes and independent clinical risk factors
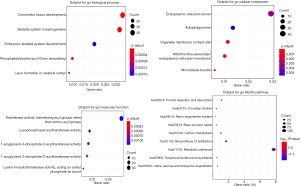
In the midnightblue module, we identified 23 key genes that met the criterion of GS >0.3, MM >0.85, and P values <0.05. These were the key genes that were highly related to DM in patients with DTC. The 23 highly connected genes in the midnightblue module were defined as the hub genes. To select the real hub genes and construct the risk-scoring model, we performed ROC analysis and AUC calculations for the hub genes. A total of three genes had AUC ≥0.75. DLX5 (AUC: 0.769), COX6B2 (AUC: 0.764), and LYPD1 (AUC: 0.760) were the most significant genes in predicting DM in DTC. Based on the results, these genes may be used as potential candidates for the clinical diagnosis of DM (Figure 4).
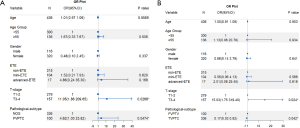
In addition, we performed univariate and multivariate analyses to explore the independent clinicopathological factors of DM in patients with DTC. Univariate analysis based on TCGA dataset revealed that T-stage (HR, 11.05; 95% CI, 1.86–209.65; and P=0.0268), and histologic subtype (HR, 4.62; 95% CI, 1.00–23.82; and P=0.0474) were significantly associated with DM. Meanwhile, T-stage (OR, 15.03; 95% CI, 1.75–319.40; P=0.024), and histologic subtype (OR, 0.17; 95% CI, 0.03–0.92; P=0.042) also served as independent predictors of DM (Figure 5).
Construction of risk score model
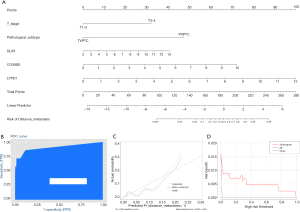
To enable clinicians to quantify the possible risk of DM in patients with DTC, a nomogram prediction model integrating the real hub genes with the independent clinicopathological risk factors was constructed (Figure 6A). This model allowed us to calculate the possibility of DM in patients with DTC by plotting a vertical line between the total points and each predictive axis. The C-index value was 0.905 and the AUC was 0.905 (95% CI, 0.876–0.926), which indicated the high discrimination of the model (Figure 6B). Calibration curves showed good agreement between the predicted risk of DM in patients with DTC and the actual status (Figure 6C). DCA revealed that the nomogram to predict DM yielded more net benefit than either the diagnose-none scheme or the diagnose-all-patients scheme (Figure 6D).
Validation the real hub genes
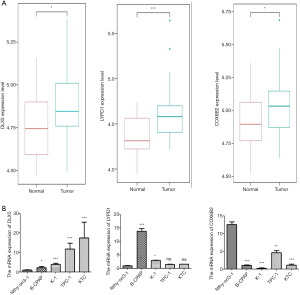
To confirm the reliability of the three real hub genes, we chose the GSE60542 database to perform difference analysis between the normal and DTC tissues. The mRNA expression levels of DLX5, COX6B2, and LYPD1 were significantly lower in the normal than DTC samples. Furthermore, qRT-PCR was performed for measuring the three real hub mRNAs expressions in Nthy-ori3-1 and in different DTC cell lines. The expression levels of DLX5 and LYPD1 were significantly lower in Nthy-ori3-1 than in the DTC cell lines, which was consistent with the mRNA expression levels in the GSE60542 database. However, the expression level of COX6B2 was contrary to the above observation (Figure 7).
Discussion
DM is a rare occurrence in thyroid cancer, and most frequently involves the bone, lung, and brain. Thyroid cancer patients who develop DM are known to have poorer prognosis than the others. Besides, more than half of these patients suffer from radioactive iodine refractory, which is associated with an even worse survival (20). Thus, early identification aggressive treatment strategy is important at a high risk for DM. Although many researchers have attempted to use next-generation sequencing and biochips to establish the key regulators of metastatic DTC (16,21), few reliable markers have been identified. Hence, the underlying molecular mechanism has not been entirely understood. WGCNA is a powerful genetic analysis tool that effectively investigates the regulatory mechanisms behind a certain characteristic and a functional interpretation for systems biology (22,23). However, little is known about the application of WGCNA in patients with DTC (24). To the best of our knowledge, this study is the first to apply the WGCNA to uncover DM-specific gene co-expression modules and identify novel hub genes. We determined key gene modules and three hub genes related to DTC progression. The findings have provided new insights into the molecular mechanism of DM in patients with DTC. The results have also yielded potential therapeutic targets for the disease.
In the present study, 9,542 DEGs were obtained from TCGA to create a co-expression network. Furthermore, 27 distinct co-expression modules were identified. The WGCNA co-expression networks revealed four gene modules that were significantly associated with DM, and the midnightblue module was most significantly positively correlated with DM (R=0.56, P=9e-06). KEGG and GO enrichment analysis indicated that the DM-related module was enriched in a variety of components, including connective tissue development, endoplasmic reticulum lumen, metabolic pathways, biosynthesis of antibiotics, and protein digestion and absorption (Figure 3). Using comprehensive analyses of MM, GS, P values, and AUC calculations, we inferred that DLX5, COX6B2, and LYPD1 were the real hub genes that were highly associated with DM in patients with DTC. Based on the findings, we suggest that DLX5, COX6B2, and LYPD1 are potential candidates for the clinical diagnosis of DM. Gandolfi et al. (25) have confirmed that the TERT promoter mutations and THYT1 genetic signature are strongly correlated with the presence of DM and reduce the survival of patients with DTC. Jung et al. (15) have reported that the PLEKHS1 promoter, a novel genetic marker, can be a useful tool in predicting the metastasis in patients with DTC. The application of the PLEKHS1 promoter signature could improve the risk stratification in the diagnostic setting of DTC patients. Unfortunately, no validated biomarkers from large and independent prospective studies are available until now. Given the complexity of metastatic DTCs and the several external and internal factors involved, it is the insufficient to explain mechanistic details and pathogenesis only from a single gene level.
Many studies have focused on the relationship between the clinicopathological characteristics and DM (4,13,14,26,27). Liu et al. (4) have evaluated 96,788 patients with DTC and revealed that sex, race, age at diagnosis, tumor size, ETE, N stage and histology subtype are significant risk factors for DM. Chen et al. (13) have reported that large tumors, ETE with follicular thyroid histology, and N1 stage have a synergistic effect on the risk for DM in patients with DTC. Our study has shown similar outcomes, identifying that histologic subtype and T-stage are independent predictors of DM. Among these independent factors, the importance of histologic subtype has been recognized in previous studies too. Follicular thyroid cancer been established to have a significantly high propensity for DM (26,28), since vascular invasion is common in this type of cancer. Additionally, tumor size also plays an important role in the development of DM. However, the current research is mainly based on the clinical parameters in predicting the metastatic DTCs, which have a low predictive accuracy.
To improve risk stratification and meet the clinical needs. we created a nomogram model. We integrated the real hub genes with the independent clinicopathological risk factors, which yielded the individual probability of DM. Based on the nomogram, we recommend aggressive therapy for patients with a high risk for DM to improve their survival, and to facilitate communication between the patients and the physicians. Substantial efforts have been taken to identify DM in patients with DTC. Kwon et al. (29) have performed conducted radiomics analysis based on grayscale ultrasound to predict DM. Nevertheless, this research used different US machines, which could have caused machine-dependent bias. Couto et al. (30) have proposed the use of a postoperative stimulated thyroglobulin (ps-Tg) value of 117.5 ng/mL as a predictor of DM in patients with DTC. However, this research ignored the influence of the remnant thyroid volume postoperatively. To confirm the reliability of the three real hub genes, we chose the GSE60542 database and different thyroid cell lines to perform difference analysis. Our findings asserted that indicate DLX5 and LYPD1 hold clinical significance for further analysis.
Undeniably, this study has a few limitations. First, it is a retrospective analysis and the number of DM patients was not adequate. Moreover, the characteristics of the enrolled patients were heterogeneous. Second, this research was based on TCGA and GEO datasets, and we did not direct obtain data from in vivo experiments. Third, we only explored the risk factors for DM. Thus, we were unable to determine the specific region of metastasis or the subsequent influence. Finally, our model was validated internally, hence, external validation should be performed to determine potential overfitting.
Conclusions
In summary, this study has resulted in the identification of a novel DM-related gene signature and has enabled the construction of a reliable predictive model for DTC patients with DM. The findings have provided an individual risk assessment tool to aid in therapeutic decision-making.
Acknowledgments
We would like to thank the TCGA and GEO databases for providing important data.
Funding: The authors appreciate the financial support provided by the National Natural Science Foundation of China (grant No. 81672885), the Natural Science Foundation of Hunan Province (grant No. 2019JJ40500) and the Fundamental Research Funds for the Central Universities of Central South University (grant No. 2020zzts259).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-383
Peer Review File: Available at https://dx.doi.org/10.21037/atm-21-383
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-383). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wang W, Yang Z, Ouyang Q. A nomogram to predict skip metastasis in papillary thyroid cancer. World J Surg Oncol 2020;18:167. [Crossref] [PubMed]
- Wang W, Zhang Z, Zhao Y, et al. Management of Lateral Multiple-Level Metastasis in N1b Papillary Thyroid Microcarcinoma. Front Oncol 2020;10:1586. [Crossref] [PubMed]
- Wang W, Meng C, Ouyang Q, et al. Magnesemia: an independent risk factor of hypocalcemia after thyroidectomy. Cancer Manag Res 2019;11:8135-44. [Crossref] [PubMed]
- Liu Z, Chen S, Huang Y, et al. Synergic effects of histology subtype, tumor size, and lymph node metastasis on distant metastasis in differentiated thyroid cancer. Ann Transl Med 2019;7:533. [Crossref] [PubMed]
- Kim H, Park SY, Jung J, et al. Improved survival after early detection of asymptomatic distant metastasis in patients with thyroid cancer. Sci Rep 2019;9:18745. [Crossref] [PubMed]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [Crossref] [PubMed]
- Tao J, Wang Y, Li L, et al. Critical Roles of ELVOL4 and IL-33 in the Progression of Obesity-Related Cardiomyopathy via Integrated Bioinformatics Analysis. Front Physiol 2020;11:542. [Crossref] [PubMed]
- Liu W, Su Y, Li S, et al. Weighted Gene Co-Expression Network Reveals Down-Regulation of Genes in Bronchopulmonary Dysplasia. Pediatr Pulmonol 2021;56:392-9. [Crossref] [PubMed]
- Pascut D, Pratama MY, Gilardi F, et al. Weighted miRNA co-expression networks analysis identifies circulating miRNA predicting overall survival in hepatocellular carcinoma patients. Sci Rep 2020;10:18967. [Crossref] [PubMed]
- Xu D, Wang Y, Wu J, et al. MTIF2 impairs 5 fluorouracil-mediated immunogenic cell death in hepatocellular carcinoma in vivo: Molecular mechanisms and therapeutic significance. Pharmacol Res 2021;163:105265 [Crossref] [PubMed]
- Chen YC, Guo YF, He H, et al. Integrative Analysis of Genomics and Transcriptome Data to Identify Potential Functional Genes of BMDs in Females. J Bone Miner Res 2016;31:1041-9. [Crossref] [PubMed]
- Xu C, Xu J, Lu L, et al. Identification of key genes and novel immune infiltration-associated biomarkers of sepsis. Innate Immun 2020;26:666-82. [Crossref] [PubMed]
- Chen D, Huang L, Chen S, et al. Innovative analysis of distant metastasis in differentiated thyroid cancer. Oncol Lett 2020;19:1985-92. [Crossref] [PubMed]
- Liu Z, Hu D, Huang Y, et al. Factors associated with distant metastasis in pediatric thyroid cancer: evaluation of the SEER database. Endocr Connect 2019;8:78-85. [Crossref] [PubMed]
- Jung CK, Jung SH, Jeon S, et al. Risk Stratification Using a Novel Genetic Classifier Including PLEKHS1 Promoter Mutations for Differentiated Thyroid Cancer with Distant Metastasis. Thyroid 2020;30:1589-600. [Crossref] [PubMed]
- Masoodi T, Siraj AK, Siraj S, et al. Whole-Exome Sequencing of Matched Primary and Metastatic Papillary Thyroid Cancer. Thyroid 2020;30:42-56. [Crossref] [PubMed]
- Landa I, Ibrahimpasic T, Boucai L, et al. Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers. J Clin Invest 2016;126:1052-66. [Crossref] [PubMed]
- Tarabichi M, Saiselet M, Tresallet C, et al. Revisiting the transcriptional analysis of primary tumours and associated nodal metastases with enhanced biological and statistical controls: application to thyroid cancer. Br J Cancer 2015;112:1665-74. [Crossref] [PubMed]
- Langfelder P, Horvath S. Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol 2007;1:54. [Crossref] [PubMed]
- Shobab L, Gomes-Lima C, Zeymo A, et al. Clinical, Pathological, and Molecular Profiling of Radioactive Iodine Refractory Differentiated Thyroid Cancer. Thyroid 2019;29:1262-8. [Crossref] [PubMed]
- Lan X, Cao J, Ye K, et al. TCR-Seq Identifies Distinct Repertoires of Distant-Metastatic and Nondistant-Metastatic Thyroid Tumors. J Clin Endocrinol Metab 2020;105:dgaa452.
- Maertens A, Tran V, Kleensang A, et al. Weighted Gene Correlation Network Analysis (WGCNA) Reveals Novel Transcription Factors Associated With Bisphenol A Dose-Response. Front Genet 2018;9:508. [Crossref] [PubMed]
- Bakhtiarizadeh MR, Hosseinpour B, Shahhoseini M, et al. Weighted Gene Co-expression Network Analysis of Endometriosis and Identification of Functional Modules Associated With Its Main Hallmarks. Front Genet 2018;9:453. [Crossref] [PubMed]
- Wang X, Fu X, Zhang J, et al. Identification and validation of m(6)A RNA methylation regulators with clinical prognostic value in Papillary thyroid cancer. Cancer Cell Int 2020;20:203. [Crossref] [PubMed]
- Gandolfi G, Ragazzi M, de Biase D, et al. Genome-wide profiling identifies the THYT1 signature as a distinctive feature of widely metastatic Papillary Thyroid Carcinomas. Oncotarget. 2017;9:1813-25. [Crossref] [PubMed]
- Khan U, Al Afif A, Aldaihani A, et al. Patient and tumor factors contributing to distant metastasis in well-differentiated thyroid cancer: a retrospective cohort study. J Otolaryngol Head Neck Surg 2020;49:78. [Crossref] [PubMed]
- Li C, Wu Q, Sun S. Radioactive Iodine Therapy in Patients With Thyroid Carcinoma With Distant Metastases: A SEER-Based Study. Cancer Control 2020;27:1073274820914661 [Crossref] [PubMed]
- Vuong HG, Duong UNP, Pham TQ, et al. Clinicopathological Risk Factors for Distant Metastasis in Differentiated Thyroid Carcinoma: A Meta-analysis. World J Surg 2018;42:1005-17. [Crossref] [PubMed]
- Kwon MR, Shin JH, Park H, et al. Radiomics Based on Thyroid Ultrasound Can Predict Distant Metastasis of Follicular Thyroid Carcinoma. J Clin Med 2020;9:2156. [Crossref] [PubMed]
- Couto JS, Almeida MFO, Trindade VCG, et al. A cutoff thyroglobulin value suggestive of distant metastases in differentiated thyroid cancer patients. Braz J Med Biol Res 2020;53:e9781 [Crossref] [PubMed]


