
Novel ETV1 mutation in small cell lung cancer transformation resistant to EGFR tyrosine kinase inhibitors
Introduction
Non-small cell lung cancer (NSCLC), which accounts for the majority of lung cancer cases, is one of the most common malignant tumors in the world (1,2). Most of the patients are in advanced stage at the time of diagnosis, and those that harbor sensitive mutations could receive targeted therapies as the standard treatment. The most common epidermal growth factor receptor (EGFR) mutations are exon 19 (Ex19del) or exon 21 (L858R) (3). The use of first-line EGFR-tyrosine kinase inhibitors (TKIs) such as gefitinib, erlotinib (first-generation), and afatinib (second-generation), which represent the standard treatment of advanced EGFR-mutated NSCLC, dramatically improves the prognosis of patients (4). However, after a median of 8–12 months of treatment, the disease will often progress, which indicates acquired resistance to EGFR-TKIs (5). The mechanisms of acquired drug resistance comprise EGFR T790M mutation, c-Met and HER2 amplification, and other driver gene mutations, as well as histological transformation to small cell lung cancer (SCLC) and an epithelial-mesenchymal (EMT) phenotype (6). Osimertinib (AZD9291) is the third generation of EGFR-TKIs, which has been shown to be effective in NSCLC patients with T790M mutation, however most patients will also inevitably develop drug resistance (7,8). Acquired EGFR C797S mutation is one of the most common osimertinib-resistant mechanisms, and SCLC transformation has also been reported (9-11).
Studies have shown that TKIs-resistant SCLC basically retains the original EGFR mutations, supporting the view that SCLC is not an independent second primary cancer, while EGFR mutations after SCLC transformation are not sensitive to EGFR-TKIs (12). Moreover, the majority of patients with histologically transformed SCLC were female non-smokers, which differed from the clinical manifestations of primary SCLC patients. These studies hypothesized that SCLC and NSCLC may have common origin cells despite their different biological and genomic characteristics (12-14). SCLC transformation is always associated with inactivating mutations in the RB1 and TP53 genes (15). Other alterations such as PTEN, CREBBP, SLIT2, EP300, and MLL mutations, as well as FGFR1 amplification have also been found in SCLC transformation cases. Although these distinctive mechanisms are well-known, a heterogeneity of the resistance to TKIs in individual is still under study and requires more attention.
Herein, we investigated two patients with EGFR-mutant lung adenocarcinoma (LUAD) who had a long-lasting response to gefitinib or osimertinib treatment and developed resistance to EGFR-TKIs due to SCLC transformation. We used a customized 639 cancer-related genes panel to analyze the genetic background of the tumor samples before EGFR-TKI treatment and after SCLC transformation, and detected novel ETV1 mutations in SCLC tissues. In vitro experiments showed that ETV1 mutations regulated achaete-scute homolog 1 (ASCL1) levels and conferred resistance to gefitinib and osimertinib, providing new insights into the role of ETV1 in lung tumorigenesis and histological transformation. This is the first report of a patient whose lung cancer transformed to SCLC after EGFR-TKI therapy with an acquired ETV1 mutations, which provides new insight into the function of the ETV1 gene and identifies a potential target that can be included in future treatment strategies for this type of cancer.
We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/atm-21-2625).
Methods
Patients and sample collection
Two NSCLC patients with EGFR sensitive mutations were recruited from the Shanghai Chest Hospital. A real-world study was conducted to detect genetic changes in the SCLC transformation after EGFR-TKI treatment. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Research Ethics Committee of Shanghai Chest Hospital (IS2118) and informed consent was taken from all the patients. Clinical information including age at diagnosis, gender, tumor histology, clinical treatment approach, outcomes, and smoking status was collected. Formalin-fixed paraffin-embedded (FFPE) tumor tissues or fresh biopsy tissues were obtained. Histological types and tumor cell contents were confirmed by two pulmonary pathologists. The efficacy assessment which is divided into complete response (CR), partial response (PR), stable disease (SD) and progressive disease (PD) was evaluated using dedicated computed tomography imaging performed and evaluated by investigator according to the Response Evaluation Criteria in Solid Tumors (RECIST 1.1).
Targeted next-generation sequencing (NGS) and bioinformatics analysis
Germline DNA (gDNA) from fresh tissues or FFPE samples de-paraffinized with xylene were extracted by using the QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Sequencing libraries were prepared using the Illumina standard library (Illumina, Inc., California, USA) according to the manufacturer’s protocols. Captured libraries were quantified by using the KAPA SYBR® FAST universal qPCR Kits (KAPA Biosystems, Boston, USA). The libraries were paired-end sequenced on an Illumina Miseq sequencer (Illumina, Hayward, CA, USA). A panel containing 639 cancer-related mutational hot genes (Singlera Genomics Inc., Shanghai, China) was used to detect multiple types of genomic variants, such as single-nucleotide variants, insertions, deletions, copy number variations, and rearrangements. The 639 involved genes are listed in Table S1.
Sequencing data were uploaded for filtering high quality reads, and clean data were aligned to The University of California at Santa Cruz (UCSC) human reference genome (GRCh37/hg19). Single-nucleotide variants, insertions or deletions, and amino acid changes were annotated by using the SnpEff (http://snpeff.sourceforge.net/). Variations detected in tumor tissues but not detected in matched blood were deemed as somatic alterations. Mutations with allele frequency >5% were defined as novel mutations. The I-TASSER server (http://zhanglab.ccmb.med.umich.edu/I-TASSER) was used to model the structure of ETV1.
Cells and reagents
The NCL-H358 cells (RRID: CVCL_1559) were maintained in RPMI1640 medium (Gibco BRL, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS, Gibco BRL, Grand Island, NY, USA) and 10 ng/mL of mouse IL3 (Cell Signaling Technology). NCL-H69 cells (RRID: CVCL_1579) were maintained in RPMI1640 medium (Gibco BRL, Grand Island, NY, USA) supplemented with 10% FBS (Gibco BRL). All cells were cultured at 37 °C in a 5% CO2-humidified atmosphere. Gefitinib and Osimertinib were purchased from Selleck Chemicals (Houston, TX, USA).
Plasmid construction
A full-length cDNA fragment of human EGFR containing activating mutations (p.L858R and p.T790M) and a full-length cDNA fragment of human ETV1 containing activating mutations (p.P159S and p.E462Q) were generated by Asia-Vector Biotechnology (Shanghai) Co. LTD. All mutated full-length EGFR or ETV1 cDNAs were induced into the PCDNA3.1 vector (Asia-Vector Biotechnology, Shanghai, China) and confirmed by direct sequencing. The sequences for mutagenesis primers are listed in Table S2.
Cell growth inhibition assay
A total of 104 transfected H358 cells expressing different EGFR-mutant variants were plated in each well of a 96-well plate, which were grown in RPMI1640 medium. Next, dimethyl sulfoxide (DMSO) or EGFR-TKIs were added at the indicated drug doses, and the cells were cultured for 72 hours. The inhibitory effects of gefitinib and osimertinib on cell growth were detected using the Cell Counting Kit-8 reagent (BBI Life Sciences, Shanghai, China). Each experiment was performed in triplicate.
Proliferation assay
A total of 104 cells were plated in each well of a 96-well plate and grown in RPMI1640 medium with 10% FBS. At the indicated times points, the cell proliferation rate was detected using the Cell Counting Kit-8 reagent (BBI Life Sciences, Shanghai, China) according to the manufacturer’s protocol. The number of viable cells was counted, and the proliferation plot was drawn using GraphPad Prism 8.0 software (GraphPad Software Inc. San Diego, CA, USA). Each experiment was performed in triplicate.
Quantitative reverse transcription PCR (qRT-PCR)
Total ribonucleic acid (RNA) was extracted using Trizol reagent (Invitrogen, CA, USA), and was then reverse transcribed into cDNA using the reverse transcriptase reagent kit (TaKaRa, Shiga, Japan). Quantitative PCR was performed using SYBR green PCR kit (TaKaRa, Shiga, Japan). Relative transcript quantities were analyzed using the ∆∆ cycle threshold (∆∆CT) method and CT values were normalized to the CT value of the GAPDH gene. Data from each experiment was then normalized to the wild-type (WT) group to detect the relative expression changes in messenger RNA (mRNA). Each experiment was performed in triplicate. The sequences of primers are listed in Table S2.
Statistical analysis
For in vitro experiments, quantitative data were presented as mean ± standard errors of the mean. A two-tailed Student t-test was performed to calculate the statistically significant differences between two groups. A P value <0.05 was considered statistically significant. For all figures: asterisk (*) means P value <0.05, (**) means P value <0.01, (***) means P value <0.001.
Results
Patient’s description
We report that two patients diagnosed with LUAD carrying EGFR activating mutations received EGFR-TKIs. Nonetheless, the patients progressed and tissue re-biopsies immediately following progression revealed a switch to SCLC histology. The clinical characteristics of the two individuals are shown in Table 1.

Full table
Case 1
A 56-year-old male, who never smoked, had intermittent hemoptysis for 5 months. Computed tomography (CT) showed an enlarged pleural mass in the inferior lobe of the right lung (4.3 cm × 2.9 cm) and multiple small nodules in bilateral lungs (Figure 1A). The pathological diagnosis was LUAD stage IV (T4N2M1), and immunohistochemical staining (IHC) confirmed the histology of pulmonary LUAD [pan cytokeratin positive (CK)+, thyroid transcription factor-1 positive (TTF1)+, cluster of differentiation 56 (CD56)−, and Ki-67 60%]. The molecular pathology results showed EGFR exon 19 deletion (2235_2249del/E746_A750del). This patient began first-line treatment with gefitinib 250 mg, once a day orally. CT scans showed a response after three months of treatment, and gefitinib was continued for a total of 12.5 months until the patient developed right pleural effusion (Figure 1A).
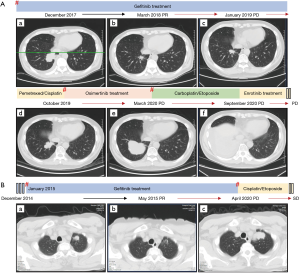
Subsequently, an EGFR T790M mutation was detected via the 639 DNA panel, and IHC showed CD56−, CK+, TTF1+, and NapsinA+ in the second biopsy specimen (Figure S1A). Osimertinib 80 mg, once daily orally, was then administered. After 4 months, osimertinib was permanently discontinued because of multiple localized pleural thickening on the right side of the lung (Figure 1A). A re-biopsy (third biopsy) was then performed, and this tumor was diagnosed as SCLC with a pathological stage of IV (T4N2M1) according to the World Health Organization (WHO) 2015 classification of lung tumors. IHC showed CD56+, CK+/−, TTF1+ (partial), NapsinA−, and synaptophysin+ (Figure S1B). Molecular analysis revealed novel genetic alterations different from those detected in previous biopsies, such as the ETV1 p.E462Q mutation. The patient then received standard carboplatin (350 mg) and etoposide (100 mg/day ×5 days) therapy. After four cycles of this combined treatment, the patient exhibited disease progression and gradual increase of pleural effusion (Figure 1A).
Case 2
A 58-year-old man with no smoking history, complained of cough, sputum, and bone pain at his primary hospital. CT showed a 1.6 cm mass in the upper lobe of the left lung, bilateral pleural thickening, and uneven bone mineral density of the left scapula and part of the vertebral body (Figure 1B). Positron emission tomography-CT (PET-CT) revealed multiple lymph node metastases in the mediastinum, left hilum, and armpit, as well as bone metastasis. A CT-guided biopsy from the primary lesion was performed and diagnosed as LUAD with a pathological stage of IV (T1aN2M1c). IHC showed CD56−, CK+, and TTF1+ (Figure S1C). EGFR L858R was positively detected by NGS, and gefitinib 250 mg, once daily orally, was administered as the first-line therapy, resulting in a partial response (PR) (Figure 1B).
After 64 months, the left pleural dissemination began to grow (Figure 1B). After hospitalization, re-biopsy was performed from the pleural tumor near the left diaphragm, and transformation to SCLC with a pathological stage of IV (T2aN2M1c) was confirmed. IHC showed CD56+, CK+, TTF1+, and NapsinA+ (Figure S1D). T790M was not detected in the tissue, however the novel ETV1 mutation (ETV1 p.P159S) was, as in the SCLC tissue analysis of patient 1. Treatment was then changed to chemotherapy (cisplatin 100 mg + etoposide 100 mg/day ×5 days), which has been used until now.
Mutational analysis of ETV1
ETV1, which is located in 7p21.2, contains 21 exons and encodes a member of the ETS (E twenty-six) family of transcription factors. ETS transcription factors can directly bind to specific DNA sequences in the promoter/enhancer regions of genes to regulate various biological processes (16). The Cancer Genome Atlas (TCGA) database showed that ETV1 mutations were most common in skin cancer, bladder urothelial cancer, nerve sheath tumor, prostate adenocarcinoma, melanoma, and endometrial carcinoma (Figure 2A). Point mutations reported in tumors were scattered in ETV1 (Figure 2B). The mutation frequency of ETV1 in LUAD and SCLC was 3.64% and 0.84%, respectively (Figure 2C,D). The mutation frequency of ETV1 reported in the Asian population with LUAD was 1.37%, while that of European and Latin American populations was 2.73% and 1.71%, respectively. The point mutations of ETV1 of LUAD were mainly concentrated in the EST-PEA3-N domain (Figure 2E).
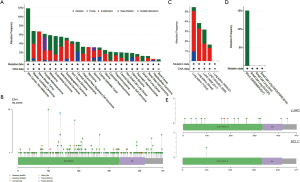
In TCGA database, only three SCLC patients were reported to have ETV1 mutation, and all of them were ETV1 S100G (Figure 2E). The ethnic origin of samples with ETV1 mutations in SCLC is unknown. Survival analysis showed that patients with ETV1 mutations had a worse overall survival (OS) trend than those without ETV1 mutations, although this was not statistically significant (Figure S2A). However, no differences were observed in progression-free survival (PFS) (Figure S2B). The worse OS trend of LUAD patients with ETV1 mutations was obvious (Figure S2C), and the disease-free survival (DFS) of these patients was significantly shortened compared to LUAD patients without ETV1 mutations (Figure S2D). SCLC patients had insufficient data to show the correlation between ETV1 mutations and survival.
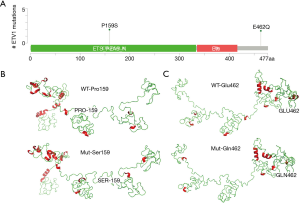
Clinical correlation analysis indicated that ETV1 mutations were significantly associated with advanced Tumor Node Metastasis (TNM) stages of LUAD (P=0.038) (Figure S3A). ETV1 mutations were more common in never-smoking LUAD patients (Figure S3B). In this study, the novel ETV1 mutations identified in two cases were ETV1 Pro159Ser (P159S), which was located in EST-PEA3-N domain, and Glu462Gln (E462Q), which was not located in a common functional domain (Figure 3A). Both of them changed the structures of the protein by computer simulation (Figure 3B,C), illustrating an important role of ETV1 in the development of lung cancer that has not been previously reported.
ETV1 mutations induced ASCL1 expression in SCLC
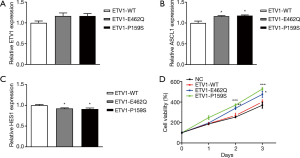
To ascertain whether ETV1 participated in neuroendocrine differentiation through regulating the expression of ASCL1, we transfected ETV1 mutant plasmids into H69 cells. We found that there was no significant difference in the expression of ETV1 mRNA (Figure 4A), while ETV1 p.E462Q and p.P159S mutant plasmids increased the ASCL1 mRNA levels after transfection (Figure 4B). We discovered that ETV1 mutations indirectly inhibited HES1 transcription (Figure 4C).
ETV1 mutations promoted SCLC proliferation in vitro
We infected H69 cells with mutant ETV1 plasmids and found that ETV1 p.E462Q and p.P159S mutations promoted SCLC proliferation in vitro. On the 2nd day after transfection, we observed that ETV1 p.E462Q and p.P159S mutations markedly induced the proliferation of tumor cells (Figure 4D).
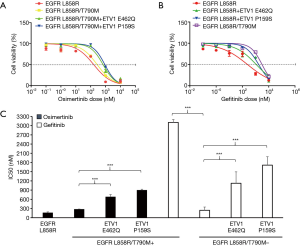
ETV1 p.E462Q and p.P159S mutations conferred resistance to gefitinib and osimertinib in vitro
We next determined whether ETV1 p.E462Q and p.P159S mutations contributed to EGFR-TKI resistance. We transfected plasmids harboring EGFR L858R or T790M (in cis) with or without ETV1 p.E462Q or p.P159S mutations into H358 cells, and exposed these cells to increased doses of osimertinib or gefitinib. As expected, cells expressing the EGFR L858R mutation were sensitive to gefitinib and osimertinib, with an IC50 of 241.8 and 160.3 nM, respectively (Figure 5A,B). Cells containing EGFR T790M were sensitive to osimertinib. EGFR mutant cells transfected with the ETV1 p.P159S mutant variant exhibited a strong resistance to osimertinib. In cells exposed to increased doses of gefitinib, the ETV1 p.E462Q and p.E462Q mutant variants exhibited a strong resistance to gefitinib when EGFR T790M was negative (Figure 5C).
Discussion
A large number of advanced NSCLC patients with EGFR mutations exhibit a good response to EGFR-TKIs. However, most patients develop resistance after an average of 8–14 months, caused by various alterations including EGFR T790M mutation, EGFR amplification, MET and HER2 amplification, and PIK3CA mutations (13). Furthermore, 5–15% of NSCLC tumors will transform into SCLC histology (13,17). Acquired resistance is the main problem that limits the clinical effect of targeted treatment with EGFR-TKIs. Although the response of transformed SCLC to chemotherapy is good, the prognosis is poor (18,19). The median survival time after confirmed transformation is 6–7 months (18). From a historical point of view, it is surprising that tumors could differentiate into NSCLC or SCLC at an early stage of tumorigenesis. EGFR mutant adenocarcinoma mainly occurs in people who never smoke, accompanied by a more sluggish natural process. Compared with NSCLC, SCLC almost always occurs in heavy smokers, and is prone to early metastasis and rapid growth (13,20).
In this study, repeated biopsies reminded us that SCLC transformed tumors might evolve from the initial adenocarcinoma rather than a concurrent cancer in the initial cases, because EGFR mutations in all cases were the same as those in the original LUAD. However, we could not rule out the possibility that SCLC with EGFR mutation existed before EGFR-TKI treatment, although we did not observe a mixed histology of NSCLC and SCLC in the tumor before treatment after carefully reviewing the histology of these two samples. However, from a clinical point of view, we suggested that these SCLCs were unlikely to exist in the early stage of tumorigenesis because classical SCLC develops very rapidly (13). RB expression loss and TP53 mutation were deemed as the main molecular mechanisms involved in predicting SCLC transformation (21). After TKIs treatment, gene alterations accumulated, and EGFR signal that stimulated NSCLC differentiation was no longer necessary for proliferation, so cells differentiated into other lineages, including EGFR mutant SCLC. In addition, alveolar type II cells can produce EGFR mutant adenocarcinoma, but after TP53 and RB1 interfere with alveolar type II cells, SCLC appears. Other alterations such as PTEN, CREBBP, SLIT2, EP300, and MLL mutations, as well as FGFR1 amplification have also been found in SCLC transformation cases (22,23). In view of these changes after EGFR-TKI exposure, resistant pluripotent stem cells subsequently differentiate into SCLC cells. Since these cells do not need EGFR signaling, they escape the influence of EGFR-TKI, so they will proliferate despite the presence of EGFR-TKI. Although there may be some EGFR mutations in transformed SCLC, the decrease or absent expression of EGFR may also lead to poor response to EGFR-TKI. The analysis of EGFR mutant tumor from adenocarcinoma to SCLC as acquired resistance mechanism showed that the lose protein expression of EGFR and the amplification level of EGFR was low. So, the response of EGFR mutant SCLC to EGFR-TKI does not seem to match those of EGFR-mutant adenocarcinoma. Thus, it is generally not recommended to detect EGFR mutations in SCLC. TKIs may not be effective in patients with SCLC due to the loss of EGFR expression at the protein level unless rare case had a positive EGFR mutation status, and they may respond to EGFR TKIs. In these two cases, ETV1 alterations were particularly related to the transformation to SCLC, which provides new insight into the function of the ETV1 gene and identifies a potential target that can be included in future treatment strategies for this type of cancer.
ETV1 is known as an oncogenic driver, and manifests as genomic translocation or amplification that causes aberrant E-twenty-six or E26 transformation-specific (ETS) expression in Ewing sarcoma, prostate cancer, and gastrointestinal stromal tumors, but not in lung cancer. In addition, ETV1 fusion has been previously described in prostate cancer, melanoma, and Ewing sarcoma (24). Recently, PTPRZ1-ETV1 and DGKB-ETV1 fusion have also been detected in glioma (24,25). Herein, we first presented two cases in which ETV1 mutations and neuroendocrine transformation exist together in the same lesion. Thus, to our knowledge, ETV1 p.E462Q and p.P159S mutations detected in these two cases have not previously been reported in lung or other tumors. The prediction of the PROVEAN software (http://provean.jcvi.org) indicated that the mutant protein had a high score of pathogenicity and was a “deleterious” mutation, so these attracted our attention.
According to the LUAD data, there was no significant difference in the mutation frequency of ETV1 between LUAD and SCLC, but there were fewer ETV1 mutant cases reported in SCLC and the mutation was single. The mutation frequency of ETV1 reported in the Asian population with LUAD was 1.37%, while that of European and Latin American populations was 2.73% and 1.71%, respectively. Only three SCLC patients with LUAD had the same ETV1 mutation. However, there was no case report of ETV1 mutations associated with SCLC type transition. All of these indicate that ETV1 mutations are rare in lung cancer, however the role of ETV1 gene in the occurrence and development of lung cancer is worth studying. In particular, computer simulation suggested that these two ETV1 mutations might cause structural changes to the ETV1 protein, which may affect the normal function of enzyme activation and DNA binding.
The maintenance of the neuroendocrine phenotype of SCLC could be detected by the presence of neuroendocrine markers, such as ASCL1. The expression of ASCL1, which was obviously present in the SCLC components while being absent or rarely present in the LUAD, highlighted the vital role of ASCL1 in small cell transformation (26). In this study, in vitro experimentation showed that ETV1 mutations increased the mRNA expression of ASCL1 in SCLC and promoted SCLC proliferation.
We also found that ETV1 mutations could confer remarkable resistance to gefitinib and osimertinib. The ETV1 p.E462Q variant exhibited mild resistance to osimertinib, while the ETV1 p.E462Q and p.P159S variants showed strong resistance to gefitinib. Previous research has demonstrated that oncogenic ETV1/4/5 are targets of Capicua (CIC), a transcription factor downstream of the receptor tyrosine kinase and mitogen-activated protein kinase pathways, which have been found to play an important role in CIC loss mediating resistance to EGFR inhibition (27). ETV1 expression is known to regulate ASCL1 expression via Notch signaling (28,29). We detected the expression levels of HES1, and suggested that ETV1 mutations regulated the expression of ASCL1 through Notch, which may be proven in further study. Ultimately, there was no significant change in the ETV1 mRNA levels after transfection of the ETV1 mutant plasmids. Collectively, these observations suggest the possibility that ETV1 mutations may represent a mechanism of intrinsic resistance to EGFR-TKIs (Figure 6). Due to the heterogeneity of tumors, it is conceivable that multiple drug resistance mechanisms may exist simultaneously in a single patient, except for SCLC transformation.

The first-line standard chemotherapy is etoposide or irinotecan combined with platinum. Concurrent or sequential radiotherapy of the thorax and mediastinum is also necessary at a limited stage. If complete remission, prophylactic cerebral irradiation (PCI) can prevent subsequent brain metastasis. In the extensive stage, chemotherapy is the main means of first-line treatment. SCLC is usually sensitive to initial treatment while most patients have recurrent disease and usually have additional metastases after initial treatment. Unfortunately, few drugs have been approved as effective second-line treatment for SCLC and the effects of them are not so good. In recent years, targeted therapy and immunotherapy have been actively tried. Nivolumab, the first FDA approved third-line treatment for SCLC, pembrolizumab, atezolizumab and durvalumab have achieved encouraging results. For other treatment options, the cytotoxic drug lurbinectedin was granted orphan drug. In addition, epigenetic inhibitors such as EZH1/2 inhibitors bring hope for the treatment of SCLC. The prospect of better treatment of SCLC lies in the combination of immunotherapy and small molecule TKI drugs. However, the success of this strategy will require the use of validated biomarkers to select the patients most likely to benefit from this strategy, including NGS for molecular alterations and a second biopsy to guide the next step of treatment and predict the prognosis.
This study highlights the importance of re-biopsy of progressive tumors in identifying a heterogeneous histopathological and genetic mechanism of resistance that might occur in EGFR-mutated NSCLCs when a rapid progression exists after an initial response. This will help clinicians to formulate the next stage of therapy, as the treatment of advanced LUAD and SCLC is completely different. In this study, the transformed SCLCs exhibited a good response to initial etoposide and cisplatin (EP) chemotherapy. Other drugs for transformed SCLC or other transformed subtypes need to be comprehensively explored in the future.
Conclusions
In this study, novel ETV1 mutations were identified in two cases of EGFR-mutant LUAD that transformed to SCLC during treatment with first- and third-generation EGFR-TKIs. ETV1 contributed to the promotion of neuroendocrine differentiation by stimulating ASCL1 expression and cell proliferation in SCLC. Moreover, ETV1 could confer resistance to gefitinib and osimertinib in vitro. In future studies, the biological function of ETV1 in lung cancer requires further exploration.
Acknowledgments
We are particularly grateful to the participants in this research.
Funding: This work was supported by the Scientific Research Project of Shanghai Municipal Health Commission (201940084), the Beijing Medical and Health Foundation (YWJKJJHKYJJ-F2213E), and the Shanghai Sailing Program (20YF1444600).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-2625
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-2625
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-2625). WYX reports that she was employed by company Singlera Genomics (Shanghai) Ltd. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Research Ethics Committee of Shanghai Chest Hospital (IS2118) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [Crossref] [PubMed]
- Bade BC, Dela Cruz CS. Lung Cancer 2020: Epidemiology, Etiology, and Prevention. Clin Chest Med 2020;41:1-24. [Crossref] [PubMed]
- Chen HF, Lei L, Wu LX, et al. Effect of icotinib on advanced lung adenocarcinoma patients with sensitive EGFR mutation detected in ctDNA by ddPCR. Transl Cancer Res 2019;8:2858-63. [Crossref]
- Novello S, Barlesi F, Califano R, et al. Metastatic non-small-cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2016;27:v1-v27. [Crossref] [PubMed]
- Jackman D, Pao W, Riely GJ, et al. Clinical definition of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. J Clin Oncol 2010;28:357-60. [Crossref] [PubMed]
- Facchinetti F, Proto C, Minari R, et al. Mechanisms of Resistance to Target Therapies in Non-small Cell Lung Cancer. Handb Exp Pharmacol 2018;249:63-89. [Crossref] [PubMed]
- Buder A, Hochmair MJ, Setinek U, et al. EGFR mutation tracking predicts survival in advanced EGFR-mutated non-small cell lung cancer patients treated with osimertinib. Transl Lung Cancer Res 2020;9:239-45. [Crossref] [PubMed]
- Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med 2017;376:629-40. [Crossref] [PubMed]
- Bordi P, Del Re M, Minari R, et al. From the beginning to resistance: Study of plasma monitoring and resistance mechanisms in a cohort of patients treated with osimertinib for advanced T790M-positive NSCLC. Lung Cancer 2019;131:78-85. [Crossref] [PubMed]
- Mu Y, Hao X, Xing P, et al. Acquired resistance to osimertinib in patients with non-small-cell lung cancer: mechanisms and clinical outcomes. J Cancer Res Clin Oncol 2020;146:2427-33. [Crossref] [PubMed]
- Ren X, Cai X, Li J, et al. Histological transformation of lung adenocarcinoma to small cell lung cancer with mutant C797S conferring acquired resistance to osimertinib. J Int Med Res 2020;48:300060520927918 [Crossref] [PubMed]
- Oser MG, Niederst MJ, Sequist LV, et al. Transformation from non-small-cell lung cancer to small-cell lung cancer: molecular drivers and cells of origin. Lancet Oncol 2015;16:e165-72. [Crossref] [PubMed]
- Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26 [Crossref] [PubMed]
- Watanabe S, Sone T, Matsui T, et al. Transformation to small-cell lung cancer following treatment with EGFR tyrosine kinase inhibitors in a patient with lung adenocarcinoma. Lung Cancer 2013;82:370-2. [Crossref] [PubMed]
- Offin M, Chan JM, Tenet M, et al. Concurrent RB1 and TP53 Alterations Define a Subset of EGFR-Mutant Lung Cancers at risk for Histologic Transformation and Inferior Clinical Outcomes. J Thorac Oncol 2019;14:1784-93. [Crossref] [PubMed]
- Sementchenko VI, Watson DK. Ets target genes: past, present and future. Oncogene 2000;19:6533-48. [Crossref] [PubMed]
- Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 2013;19:2240-7. [Crossref] [PubMed]
- Roca E, Gurizzan C, Amoroso V, et al. Outcome of patients with lung adenocarcinoma with transformation to small-cell lung cancer following tyrosine kinase inhibitors treatment: A systematic review and pooled analysis. Cancer Treat Rev 2017;59:117-22. [Crossref] [PubMed]
- Ma S, He Z, Fu H, et al. Dynamic changes of acquired T790M mutation and small cell lung cancer transformation in a patient with EGFR-mutant adenocarcinoma after first- and third-generation EGFR-TKIs: a case report. Transl Lung Cancer Res 2020;9:139-43. [Crossref] [PubMed]
- Basumallik N, Agarwal M. Small Cell Lung Cancer. StatPearls. Treasure Island, FL; 2020.
- Lee JK, Lee J, Kim S, et al. Clonal History and Genetic Predictors of Transformation Into Small-Cell Carcinomas From Lung Adenocarcinomas. J Clin Oncol 2017;35:3065-74. [Crossref] [PubMed]
- Forgacs E, Biesterveld EJ, Sekido Y, et al. Mutation analysis of the PTEN/MMAC1 gene in lung cancer. Oncogene 1998;17:1557-65. [Crossref] [PubMed]
- Peifer M, Fernández-Cuesta L, Sos ML, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 2012;44:1104-10. [Crossref] [PubMed]
- Johnson A, Severson E, Gay L, et al. Comprehensive Genomic Profiling of 282 Pediatric Low- and High-Grade Gliomas Reveals Genomic Drivers, Tumor Mutational Burden, and Hypermutation Signatures. Oncologist 2017;22:1478-90. [Crossref] [PubMed]
- Matjasic A, Zupan A, Bostjancic E, et al. A novel PTPRZ1-ETV1 fusion in gliomas. Brain Pathol 2020;30:226-34. [Crossref] [PubMed]
- Lin MW, Su KY, Su TJ, et al. Clinicopathological and genomic comparisons between different histologic components in combined small cell lung cancer and non-small cell lung cancer. Lung Cancer 2018;125:282-90. [Crossref] [PubMed]
- Liao S, Davoli T, Leng Y, et al. A genetic interaction analysis identifies cancer drivers that modify EGFR dependency. Genes Dev 2017;31:184-96. [Crossref] [PubMed]
- Akazawa C, Sasai Y, Nakanishi S, et al. Molecular characterization of a rat negative regulator with a basic helix-loop-helix structure predominantly expressed in the developing nervous system. J Biol Chem 1992;267:21879-85. [Crossref] [PubMed]
- Kovach C, Dixit R, Li S, et al. Neurog2 simultaneously activates and represses alternative gene expression programs in the developing neocortex. Cereb Cortex 2013;23:1884-900. [Crossref] [PubMed]
(English Language Editor: A. Kassem)


