
Role of competitive endogenous RNA networks in the pathogenesis of coronary artery disease
Introduction
Coronary artery disease (CAD) is a commonly occurring type of cardiovascular disease. It can result in angina pectoris, myocardial infarction, heart failure, and arrhythmia. In certain cases, the occurrence of atherosclerosis results in coronary artery stenosis and insufficient blood supply to the myocardium, leading to death. Current treatments for CAD include percutaneous coronary intervention, drug therapy, and coronary artery bypass grafting. The age and sex standardized incidence of CAD was 436 per 100,000 in 2015 (1). Although patient quality of life has improved through advancements in medical treatment and secondary prevention, it still remains that 35% of CAD patients suffer relapse (2).
Recent studies have suggested new molecules involved in the progression of CAD. For example, ANRIL is a long non-coding RNA (lncRNA) expressed at low levels in the serum of CAD patients, and high expression of ANRIL predicts poor prognosis in CAD patients (3). microRNA (miR)-128 negatively regulates the expression of IRS1, which promotes the viability and migration of rat cardiac microvascular endothelial cells and inhibits cell apoptosis (4). The identification of new molecules involved in CAD progression is essential to better understand its pathogenesis and to provide new targets for the treatment of CAD.
A recent study showed that competitive endogenous RNA (ceRNA) regulation networks play an important role in heart diseases. For example, an endogenous competitive relationship between the lncRNA MEG3 and miR-145 was identified. The overexpression of MEG3 decreased the expression of miR-145, which in turn increased the expression of the target gene PDCD4 and promoted cardiomyocyte apoptosis (5). Recently, despite a ceRNA literature report on CAD, it revealed 11 pathways and 15 key genes related to CAD, which provided options for the treatment of CAD (6). However, our study used different datasets from the previous ones. These datasets correspond to lncRNA, miRNA and mRNA chip analysis results respectively, so that we can integrate and analyze data from a wider dimension. From another new perspective, we constructed a ceRNA network to reveal the molecular mechanisms related to CAD, and combined the results reported in previous articles to provide some more comprehensive and higher-quality choices for the target treatment of CAD.
We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/atm-21-2737).
Methods
CAD data
The lncRNA, miRNA, and mRNA expression profiles were downloaded from GEO (https://www.ncbi.nlm.nih.gov/geo/). These expression profiles were from plasma samples of patients with CAD. The lncRNA microarray data were obtained from the GSE68506 (comprising three CAD patients and three normal controls). The miRNA expression data were obtained from the GSE59421 (comprising 33 CAD patients and 63 normal controls), and the mRNA expression data were obtained from the GSE20129 (comprising 48 CAD patients and 71 normal controls). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Identification of differentially expressed lncRNAs (DElncRNAs), miRNAs (DEmiRNAs), and mRNAs (DEmRNAs)
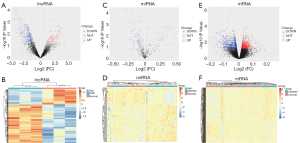
The Bioconductor Limma (7) package and Perl were used to identify DElncRNAs, DEmiRNAs, and DEmRNAs in the CAD patients and normal controls. DElncRNAs, DEmiRNAs, and DEmRNAs were screened by thresholds of P<0.05. After the DE analysis (Figure 1), we visualized the DElncRNAs, DEmiRNAs, and DEmRNAs between CAD patients and normal controls. Clustering heat maps and volcano maps were made using the R package “pheatmap”.
Construction of the ceRNA network
To better comprehend the relationships between the DE mRNAs, miRNAs, and lncRNAs, the lncRNA-mediated ceRNA network of CAD was constructed as follows. First, we used the miRCode database (http://www.mircode.org/) (8) to predict relationships between the lncRNAs and miRNAs. Next, the miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/), miRDB (http://www.mirdb.org/), and TargetScan (http://www.targetscan.org/) (9-11) databases were used to obtain the miRNA-targeted mRNAs. To improve the effectiveness of our results, we showed miRNA-targeted mRNA both in the miRTarBase, miRDB, and TargetScan databases to establish a lncRNA-miRNA-mRNA network. Finally, Cytoscape (http://www.cytoscape.org/) 3.8.1 (12) software was used to visualize the results.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG)
GO is a popular bioinformatics tool used to analyze the biological functions involved in target genes (13,14). The KEGG is a large-scale molecular dataset, generated using high-throughput experimental methods, that is used to understand the biological signaling pathways involved in genes (15). In the Database for Annotation, Visualization and Integrated Discovery (DAVID; https://david.ncifcrf.gov/) (16), we used GO annotations and KEGG to analyze the biological functions and signaling mechanisms involved in DEmRNAs. P<0.05 was considered statistically significant.
Construction of protein-protein interaction (PPI) network and identification of key genes
The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) (http://string-db.org/) (17) was used to predict the PPI network based on the gene symbols (18).The PPI network of the DEmRNAs was constructed using the STRING database, using a combination score of >0.4, and the differences were statistically significant. Next, we visualized the molecular interaction network using Cytoscape. The key genes with the highest scores were screened out using the maximum clique centrality (MCC) method in the cytoHubba plug-in of Cytoscape (19).
Statistical analysis
All statistical analyses were performed using R (v.4.0.3) software, Perl (v.5.28.1) and GraphPad Prism 9 software. The P value <0.05 was considered statistically significant.
Results
Identification of DElncRNAs, DEmiRNAs, and DEmRNAs
Based on the screening criterion of P<0.05, a total of 264 DElncRNAs (179 downregulated and 85 upregulated lncRNAs), 106 DEmiRNAs (73 downregulated and 33 upregulated miRNAs), and 1,879 DEmRNAs (1,066 downregulated and 813 upregulated mRNAs) were identified between the CAD and normal control groups. Heatmap clustering indicated that the DElncRNAs, DEmiRNAs, and DEmRNAs had clearly defined differences in expression between the two groups (Figure 2).
Biological functions and signaling mechanisms related to the DEmRNAs
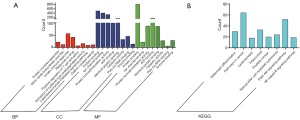
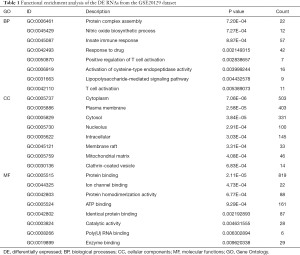
Through the GO annotations, we found that the DEmRNAs in the GSE20129 were enriched in protein complex assembly, nitric oxide biosynthesis, innate immune response, cytoplasm, plasma membrane, cytosol, etc. (Figure 3A, Table 1). The KEGG pathway analysis showed that the DEmRNAs in the GSE20129 were mainly involved in the PI3K-Akt signaling pathway, tuberculosis, cancer pathways, etc. (Figure 3B, Table 2).
Full table

Full table
ceRNA network
Among the DElncRNAs, DEmiRNAs, and DEmRNAs, 21 lncRNAs (18 downregulated and 3 upregulated), 13 miRNAs (13 downregulated), and 143 mRNAs (86 downregulated and 57 upregulated) were involved in the proposed ceRNA network (Figure 4, Table 3).
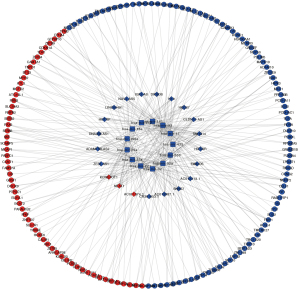
Full table
GO and KEGG analysis of the ceRNA network
DEmRNAs in the ceRNA network were enriched in protein phosphorylation, drug responses, viral processes, cytoplasm, nucleus, cytosol, protein binding, ATP binding, protein serine kinase activity, etc. (Figure 5A-5C, Table 4). The KEGG pathway analysis showed that the DEmRNAs in the ceRNA network were involved in the PI3K-Akt signaling pathway, neurotrophin signaling pathway, cancer pathways, etc. (Figure 5D, Table 5).
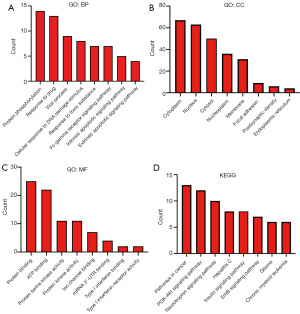
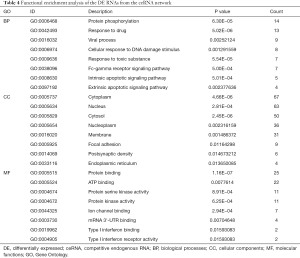
Full table

Full table
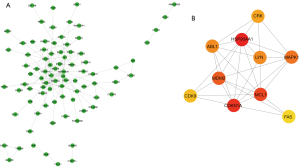
Key genes in the PPI network
The PPI network was constructed based on STRING in Cytoscape (Figure 6A). The MCC method from the cytoHubba app in Cytoscape was used to screen for genes with higher scores, which were considered key genes. The top 10 key genes were HSP90AA1, CDKN1A, MCL1, MDM2, MAPK1, ABL1, LYN, CRK, CDK9, and FAS (Figure 6B, Table 6).

Full table
Discussion
Because of its high risk for emergencies, CAD is the leading disease-related cause of human death (20,21). The World Health Organization estimates that 7.4 million people die of CAD every year (22). Although some progress has been made in the diagnosis and treatment of CAD, its molecular mechanisms are still unclear. Therefore, there is a pressing need for further research to identify potential targets for CAD treatment.
The focus of this study was to screen lncRNA, miRNA and mRNA differential genes related to CAD through GEO database, and then construct lncRNA-miRNA-mRNA network. Finally, 10 key genes and some signaling pathways were identified, which provided a better entry point for the basic research on the pathological mechanism of CAD in the future.
Increasing evidence indicates that lncRNAs can competitively bind miRNAs through sponge adsorption to modulate cell proliferation, metastasis, differentiation, and apoptosis to regulate the initiation and progression of diseases (23). For example, the lncRNA KCNQ1OT1 mediates miR-466i-5p downregulation, inducing high expression of the target gene Tead1 and leading to cardiomyocyte damage (24). In addition, the lncRNAs SNHG14 and SNHG7 competitively sponge miR-322-5p and miR-34-5p, respectively, increasing the expression levels of PCDH17 and ROCK1, leading to cardiomyocyte hypertrophy and fibrosis (25,26).
Downregulation of downstream miR-125a-5p via the lncRNA NEAT1 leads to overexpression of the target gene BCL2L12 and results in cardiomyocyte apoptosis (27). The lncRNA HOTAIR downregulates miR-545 to increase the expression of EGFR and p-ERK, significantly improving cardiomyocyte activity and inhibiting cell apoptosis (28). Thus, these lncRNAs from the ceRNA network play important roles in CAD, aging, and apoptosis. In this study, a lncRNA-miRNA-mRNA ceRNA network was constructed through bioinformatics to identify candidate molecules for the treatment of CAD.
We found the key genes in the constructed PPI network to be HSP90AA1, CDKN1A, MCL1, MDM2, MAPK1, ABL1, LYN, CRK, CDK9, and FAS. HSP90AA1 is the most extensively studied member of the heat shock protein (Hsp) family, whose main role is to maintain protein homeostasis and cell protection. HSP90AA1 overexpression reduced the apoptosis of neonatal rat ventricular cells induced by oxygen glucose deprivation (29). CDKN1A encodes a potent cyclin-dependent kinase inhibitor, which plays a crucial regulative role in cell-cycle progression. Knockdown of CDKN1A can inhibit cardiomyocyte hypertrophy and fibrosis while protecting myocardium in mice (30). MCL1 encodes an anti-apoptotic protein, which is a member of the Bcl-2 family. Knockout of MCL1 gene can cause mitochondrial dysfunction, which impairs the development of autophagy and heart failure (31). In the mouse model of atherosclerosis, the combination of lncRNA-p21 and MDM2 leads to the proliferation of vascular smooth muscle cells (VSMCs), reduces the apoptosis of VSMCs, and participates in the pathogenesis of atherosclerosis (32).
MAPK1 is a protein-coding gene with transferase activity and tyrosine kinase activity that is involved in the transfer of phosphorus-containing groups in signaling pathways. Studies have shown that MAPK1 is upregulated by miR-140-3p and inhibits CAD cell apoptosis (33). Knockout of ABL1 gene inhibits c-Abl activity and significantly reduces apoptosis of VSMCs and synthetic phenotypic transformation induced by Ang II both in vivo and in vitro (34). CRK plays a key role in Rac1-induced membrane ruffling and Rap1-mediated nascent focal complex stabilization, which contributed to ephrin-B1-induced human aortic endothelial cells migration (35).CDK9 has been shown to regulate cardiomyocyte hypertrophy, and recent evidence suggests that it is involved in cardiomyocyte proliferation (36). FAS encoded by this gene is a member of the TNF-receptor superfamily, which contains a death domain. Fas and FasL show interdependence with inflammatory markers in the process of apoptosis in patients with ischemic heart disease (37).
The PI3K-Akt signaling pathway is aberrantly activated during the progression of heart disease. Overexpression of IGF-1 can activate the PI3K-Akt pathway, inducing physiological myocardial hypertrophy and myocardial infarction (38). The NF-κB signaling pathway is also frequently involved in the pathogenesis of heart diseases. For example, miR-21 protects cardiomyocytes from apoptosis that is induced by palmitate through the caspase-3/NF-κB signal pathways (39). Consistent with these results, we found that the PI3K-Akt signaling pathway and the NF-κB signaling pathway were enriched by the DEmRNAs, suggesting that these pathways play an important role in the pathology of CAD. However, further tissue and cell studies still need to be carried out to validate the expression differences of the predicted key genes and determine their roles in the relevant pathways.
Conclusions
The ceRNA network constructed in this study identified new candidate molecules involved in the pathogenesis of CAD and may lead to improved treatment of CAD patients.
Acknowledgments
Funding: Scientific research start-up fund for young and middle-aged outstanding talents.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-2737
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-2737). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sundaram V, Bloom C, Zakeri R, et al. Temporal trends in the incidence, treatment patterns, and outcomes of coronary artery disease and peripheral artery disease in the UK, 2006-2015. Eur Heart J 2020;41:1636-49. [Crossref] [PubMed]
- Zhang HW, Jin JL, Cao YX, et al. Heart-type fatty acid binding protein predicts cardiovascular events in patients with stable coronary artery disease: a prospective cohort study. Ann Transl Med 2020;8:1349. [Crossref] [PubMed]
- Liu ZF, Hu WW, Li R, et al. Expression of lncRNA-ANRIL in patients with coronary heart disease before and after treatment and its short-term prognosis predictive value. Eur Rev Med Pharmacol Sci 2020;24:376-84. [PubMed]
- Yan P, Sun C, Ma J, et al. MicroRNA-128 confers protection against cardiac microvascular endothelial cell injury in coronary heart disease via negative regulation of IRS1. J Cell Physiol 2019;234:13452-63. [Crossref] [PubMed]
- Chen Y, Zhang Z, Zhu D, et al. Long non-coding RNA MEG3 serves as a ceRNA for microRNA-145 to induce apoptosis of AC16 cardiomyocytes under high glucose condition. Biosci Rep 2019;39:BSR20190444 [Crossref] [PubMed]
- Kang S, Ye Y, Xia G, et al. Coronary artery disease: differential expression of ceRNAs and interaction analyses. Ann Transl Med 2021;9:229. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47 [Crossref] [PubMed]
- Jeggari A, Marks DS, Larsson E. miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics 2012;28:2062-3. [Crossref] [PubMed]
- Chou CH, Chang NW, Shrestha S, et al. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res 2016;44:D239-47. [Crossref] [PubMed]
- Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 2015;43:D146-52. [Crossref] [PubMed]
- Fromm B, Billipp T, Peck LE, et al. A uniform system for the annotation of vertebrate microRNA genes and the evolution of the human microRNAome. Annu Rev Genet 2015;49:213-42. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25-9. [Crossref] [PubMed]
- Guo Q, Ke XX, Liu Z, et al. Evaluation of the prognostic value of STEAP1 in lung adenocarcinoma and insights into its potential molecular pathways via bioinformatic analysis. Front Genet 2020;11:242. [Crossref] [PubMed]
- Kanehisa M, Furumichi M, Tanabe M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017;45:D353-61. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 2013;41:D808-15. [Crossref] [PubMed]
- Yang L, Zhao X, Tang X. Predicting disease-related proteins based on clique backbone in protein-protein interaction network. Int J Biol Sci 2014;10:677-88. [Crossref] [PubMed]
- Smoot ME, Ono K, Ruscheinski J, et al. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 2011;27:431-2. [Crossref] [PubMed]
- Moran AE, Roth GA, Narula J, et al. 1990-2010 global cardiovascular disease atlas. Glob Heart 2014;9:3-16. [Crossref] [PubMed]
- Moran AE, Wood DA, Narula J. The 2000-2016 WHF global atlas of CVD: take two. Glob Heart 2018;13:139-41. [Crossref] [PubMed]
- Thomas H, Diamond J, Vieco A, et al. Global atlas of cardiovascular disease 2000-2016: the path to prevention and control. Glob Heart 2018;13:143-63. [Crossref] [PubMed]
- Huang X, Zhou X, Hu Q, et al. Advances in esophageal cancer: a new perspective on pathogenesis associated with long non-coding RNAs. Cancer Lett 2018;413:94-101. [Crossref] [PubMed]
- Liao B, Dong S, Xu Z, et al. LncRNA Kcnq1ot1 renders cardiomyocytes apoptosis in acute myocardial infarction model by up-regulating Tead1. Life Sci 2020;256:117811 [Crossref] [PubMed]
- Long Y, Wang L, Li Z. SP1-induced SNHG14 aggravates hypertrophic response in in vitro model of cardiac hypertrophy via up-regulation of PCDH17. J Cell Mol Med 2020;24:7115-26. [Crossref] [PubMed]
- Wang J, Zhang S, Li X, et al. LncRNA SNHG7 promotes cardiac remodeling by upregulating ROCK1 via sponging miR-34-5p. Aging (Albany NY) 2020;12:10441-56. [Crossref] [PubMed]
- Yan H, Liang H, Liu L, et al. Long noncoding RNA NEAT1 sponges miR-125a-5p to suppress cardiomyocyte apoptosis via BCL2L12. Mol Med Rep 2019;19:4468-74. [Crossref] [PubMed]
- Li Y, Zhao W, Shi R, et al. Rs4759314 polymorphism located in HOTAIR is associated with the risk of congenital heart disease by alternating downstream signaling via reducing its expression. J Cell Biochem 2018;119:8112-22. [Crossref] [PubMed]
- Zhu WS, Guo W, Zhu JN, et al. Hsp90aa1: a novel target gene of miR-1 in cardiac ischemia/reperfusion injury. Sci Rep 2016;6:24498. [Crossref] [PubMed]
- Xu M, Tang Q. Cdkn1α knockout alleviates pressure overload induced cardiac remodeling. J Am Coll Cardiol 2016;68:C58. [Crossref]
- Cai CL, Jin L, Lang XL, et al. Long noncoding RNA XIST regulates cardiomyocyte apoptosis by targeting miR-873-5p/MCL1 axis. Eur Rev Med Pharmacol Sci 2020;24:12878-86. [PubMed]
- Han Y, Cai J, Chen C, et al. LincRNA-p21 feedback enhances P53 activity via interaction with MDM2 function in vascular smooth muscle cells proliferation dominantly neointimal hyperplasia of atherosclerosis. Cardiology (Switzerland) 2013;126:40-1.
- Zhang H, Ji N, Gong X, et al. NEAT1/miR-140-3p/MAPK1 mediates the viability and survival of coronary endothelial cells and affects coronary atherosclerotic heart disease. Acta Biochim Biophys Sin (Shanghai) 2020;52:967-74. [Crossref] [PubMed]
- Zhou X, Cheng J, Chen Z, et al. Role of c-Abl in Ang II-induced aortic dissection formation: Potential regulatory efficacy on phenotypic transformation and apoptosis of VSMCs. Life Sci 2020;256:117882 [Crossref] [PubMed]
- Nagashima K, Endo A, Ogita H, et al. Adaptor protein Crk is required for ephrin-B1-induced membrane ruffling and focal complex assembly of human aortic endothelial cells. Mol Biol Cell 2002;13:4231-42. [Crossref] [PubMed]
- Matrone G, Wilson KS, Maqsood S, et al. CDK9 and its repressor LARP7 modulate cardiomyocyte proliferation and response to injury in the zebrafish heart. J Cell Sci 2015;128:4560-71. [Crossref] [PubMed]
- Ristic T, Cosic V, Deljanin Ilic M, et al. Interdependence between serum Fas/FasL levels and inflammatory markers in patients with ischemic heart disease. Clin Chem Lab Med 2015;53:S463.
- McMullen JR, Shioi T, Huang WY, et al. The insulin-like growth factor 1 receptor induces physiological heart growth via the phosphoinositide 3-kinase(p110alpha) pathway. J Biol Chem 2004;279:4782-93. [Crossref] [PubMed]
- Zhou X, Chang B, Gu Y. MicroRNA-21 abrogates palmitate-induced cardiomyocyte apoptosis through caspase-3/NF-κB signal pathways. Anatol J Cardiol 2018;20:336-46. [PubMed]
(English Language Editor: K. Brown)


