
The gut microbiome in microscopic polyangiitis with kidney involvement: common and unique alterations, clinical association and values for disease diagnosis and outcome prediction
Introduction
Anti-neutrophil cytoplasmic antibody (ANCA) associated vasculitis is an autoimmune disorder characterized by multi-systemic involvement (1). The annual incidence is estimated to 21.8 and 22.6 per million in the United Kingdoms and Japan, respectively (2). Microscopic polyangiitis (MPA) and granulomatosis with polyangiitis (GPA) are two major phenotypes with evident differences in clinical manifestation, pathological features and geographical distribution (3,4). In China, MPA is more common than GPA (5). Notably, more than 90% of MPA patients suffer from kidney involvement (6). In addition, kidney involvement is associated with increased mortality, and advanced renal injury at diagnosis can predict poor renal prognosis (7).
Gut bacteria are essential for the homeostasis of mucosal immunity and the integrity of intestinal barrier (8). Disturbed gut community has been linked to many diseases, including type 2 diabetes (9), atherosclerotic cardiovascular disease (10,11), hepatocellular carcinoma (12), systemic lupus erythematosus (SLE) (13) and chronic kidney disease (CKD) (14-16). Accumulating evidence suggests that certain bacteria may be implicated in the development of ANCA associated vasculitis. Firstly, GPA patients usually have upper respiratory tract involvement and are frequently affected by infective events during relapse (17). Secondly, chronic nasal carriage of Staphylococcus Aureus in GPA correlates with increased frequency of relapse and maintenance of trimethoprim-sulfamethoxazole seems useful in patients with upper airway-limited GPA and history of frequent relapses (18,19). In mechanism, studies showed that two bacteria-derived peptides could trigger autoimmunity to autoantigen for ANCA and induce glomerulonephritis in mice (20,21). However, the characteristics of the gut microbiome of MPA have not been investigated, and its role in the development of MPA remains unknown.
Renal biopsy is valuable for the diagnosis and evaluation of MPA related renal injury. However, due to its invasive characteristics, its application is greatly limited. ANCA test is helpful for the diagnosis of MPA, but ANCA is negative in about 15% of patients (22). Thus, more noninvasive tools are urgently needed for the diagnosis and evaluation of MPA. Intriguingly, with advances on genetic sequencing and artificial intelligence, the gut microbiome has been investigated, and its changes were closely related to various diseases, such as hepatocellular carcinoma (12), juvenile idiopathic arthritis and SLE (13).
In this cross-sectional study, we performed 16S rRNA amplicon sequencing to profile the gut microbiome of MPA patients, and further explored its connection with clinical indices, as well as its potential as a diagnosis tool applying machine-learning methods. We present the following article in accordance with the MDAR and STROBE reporting checklist (available at https://dx.doi.org/10.21037/atm-21-1315).
Methods
Study population and sample collection
Patients with incipient active MPA (aMPA), inactive MPA (inMPA), and age and gender-matched healthy controls (HCs) were recruited from 2017 to 2019 at the first affiliated hospital, Zhejiang University School of Medicine. MPA was diagnosed according to 2012 International Chapel Hill Consensus Conference on the Nomenclature of Vasculitis (23). To minimize the heterogeneity of population, we enrolled only myeloperoxidase (MPO)-ANCA positive patients with kidney involvement. Patients with acute infection, diabetes mellitus, obesity, cancer and other autoimmune disorders (such as inflammatory bowel disease, rheumatoid arthritis and SLE), as well as those who took probiotics or antibiotics within one month before enrollment and at sampling were excluded.
The demographic and laboratory data were obtained through electronic medical record system. Birmingham Vasculitis Activity Score (BVAS) (24) was used to assess MPA activity, and BVAS =0 indicated remission of disease. The patients in aMPA cohort were prospectively followed up for 6 to 12 months. Events of death or end-stage renal disease (ESRD) were recorded.
For the aMPA cohort, fresh fecal samples were collected during in-patient care. For the inMPA and HC cohorts, the samples were collected at clinical visit or during health examination. All samples were frozen at −80 °C within one hour after collection. Our study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the ethics committee of the First Affiliated Hospital, Zhejiang University School of Medicine (approval No. 2017-694). Informed consents were available from all participants before sampling.
DNA extraction, library preparation and sequencing
Total genomic DNA was extracted from all fecal samples using the PowerSoil® DNA Isolation Kit (MO BIO, USA) following the manufacturer’s instructions. PCR was then performed to amplify the V3-V4 variable region of 16S rRNA gene. The 16S amplicon library was constructed using the TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina, USA) and sequenced on an Ion S5TM platform at Zhejiang Institute of Microbiology (Hangzhou, China).
Sequence processing
All sequencing reads were filtered by the Quantitative Insights Into Microbial Ecology (QIIME, version 1.9.1) (25) and then aligned with GOLD database by UCHIME algorithm to eliminate chimera sequences (26). The obtained reads were subsequently clustered into different Operational Taxonomic Units (OTUs) according to similarity greater than 97% by Uparse 8.1 (27). Sequences with the highest frequency in each OTU were screened as representative sequences. For further taxonomic annotation, representative sequences were aligned with SILVA reference database (28) by uclust (29) at 90% threshold.
Analysis of microbial diversity and comparison of taxonomic abundance
α-diversity including bacterial richness (Ace and Chao1) and diversity (Shannon and Simpson) were calculated by QIIME. Non-metric multi-dimensional Scaling (NMDS), a typical β-diversity analysis, was performed by vegan package. Analysis of similarity (Anosim) was further conducted to statistically compare the community structure among the three groups. Relative abundance of phyla and genera greater than 0.01% were compared between any two groups using Wilcoxon rank sum test with “Benjamini-Hochberg” adjusted P values.
Correlation between altered genera and clinical indices, differential analysis on OTUs
Spearman correlations between the abundance of differential genera and clinical indices in MPA patients were calculated using the psych package 1.9.12. The obtained matrix with “Holm” adjusted P values was visualized by corrplot package 0.84. In order to find OTU markers for severe kidney damage and predicting renal prognosis, we compared the OTU abundance in aMPA patients based on their initial status on dialysis and final status on kidney survival. This differential analysis was performed using ALDEx2 package 1.18.0 and visualized by ggplot2 package 3.3.1.
Microbial community-based classifier models
In order to evaluate the feasibility of gut microbiome in assisting diagnosis of MPA, we implemented a 10-fold cross-validation approach on random forest models (randomForest package 4.6-14) (30) using the profile of OTU abundance from aMPA and HCs. For predicting disease activity, the same method was adopted using the profile of OTU abundance from aMPA and inMPA. The error curves of 5 trials of 10-fold cross-validation were averaged. The minimum averaged error plus the corresponding standard deviation (SD) was set as the cutoff. The optimal set of OTU markers was selected based on two terms: cross-validation error less than the cutoff and the least OTU numbers. Next, the probability of MPA or active disease was compared between groups using the optimal set of OTUs. The ROC curve was drawn using pROC package 1.16.1.
Statistical analysis
For continuous variables, normally distributed data was expressed as mean ± SD and skewed distributed data was expressed as median with interquartile range (IQR). Categorical variables were expressed as percentage. Difference between two groups was compared by Wilcoxon rank sum test or unpaired t-test. Multiple group comparisons were conducted by Kruskal-Wallis test and P values were adjusted using “Benjamini-Hochberg” method or “Holm” method. A difference with P<0.05 was considered a statistically significance. All statistical analysis was performed using R 3.6.1 or SPSS 22.0.
Results
Clinical and laboratory characteristics of all participants
A total of 71 different patients with MPA (35 with active MPA and 36 with inactive MPA) and 34 HCs met our inclusion criteria (Figure S1). The clinical, laboratory and histopathological findings of all participants were detailed in Table S1. There was no significant difference in age, gender and BMI among these groups (Table 1). Both aMPA and inMPA patients had higher levels of white blood cell (P<0.001, 0.001, respectively) and serum creatinine (both P values <0.001), and lower hemoglobin level (both P values <0.001) compared with HCs. aMPA patients had higher levels of C-reactive protein, erythrocyte sedimentation rate, MPO-ANCA titre, serum creatinine, urine protein and BVAS compared with inMPA patients (all P values <0.001 except 0.004 for MPO-ANCA titre). Ten out of 35 aMPA patients had severe renal injury requiring dialysis during the initial hospitalization. For immunosuppressive treatment, the proportion of patients receiving steroid therapy was similar in aMPA group and inMPA group, while more inMPA patients received mycophenolate mofetil (MMF) compared with aMPA patients (64% versus 9%, P<0.01). At the final follow-up, 18 out of 34 aMPA patients progressed to ESRD, and one aMPA patient died of respiratory and renal failure during hospitalization.
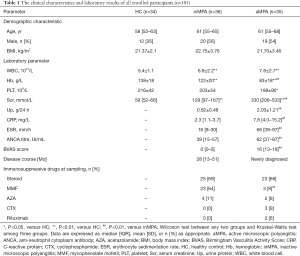
Full table
Richness and diversity of microbial community
The sequencing data from 105 fecal samples were clustered into 1,754 different OTUs, of which 40% were shared by the three groups (Figure 1A). The community richness assessed by Ace and Chao1 index showed no significant difference among the groups, except that the Chao1 index of the aMPA group was lower than that of the inMPA group (Figure 1B,1C). The community diversity assessed by Simpson and Shannon index was lowest in the aMPA group and highest in the HCs (Figure 1D,1E). In addition, distinct microbial composition among the groups was initially illustrated by NMDS plot (Figure 1F, stress =0.18) and further confirmed by pairwise comparisons in ANOSIM (all P values =0.001 between any two groups).

Composition of microflora and differential taxa
After removing unassigned OTUs, we annotated the remaining 1,562 OTUs into 24 phyla, 37 classes, 86 orders, 149 families and 354 genera. At phylum level, the most common taxa were phylum Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, Fusobacteria (Figure 2A, Figure S2). At genus level, the most common taxa were genus Bacteroides, Faecalibacterium, Blautia, Streptococcus, Escherichia Shigella (Figure 2B, Figure S3).
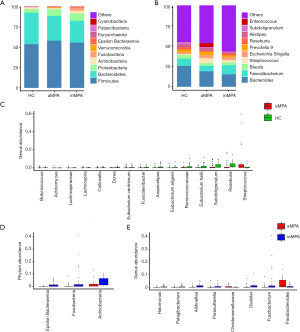
Next, the abundance of phyla or genera greater than 0.01% was compared between each pair of groups. Three phyla—Actinobacteria, Fusobacteria and Epsilonbacteraeota, were significantly higher in the inMPA group than those in the HCs group (Figure S4A, Table S2, adjusted P<0.001, 0.003, <0.001, respectively). However, there was no statistical difference of phylum abundance between the aMPA group and the HCs group. At genus level, two genera including Actinomyces and Streptococcus were more abundant in the aMPA group (adjusted P=0.001 and 0.04) and the inMPA group (adjusted P<0.001, 0.002) than those in HCs group, while six genera including Subdoligranulum, Eubacterium hallii, Ruminococcaceae UCG013, Eubacterium ventriosum, Dorea and Butyricicoccus were more abundant in the HCs group than those in the aMPA group (adjusted P=0.01, 0.02, 0.01, 0.02, 0.007 and 0.004, respectively) and the inMPA group (adjusted P=0.002, 0.001, <0.001, <0.001, 0.008 and 0.001, respectively) (Figure 2C, Figure S4B, Table S3). Among the above six decreased genera in the MPA groups, Subdoligranulum, Ruminococcaceae UCG013 and Butyricicoccus belong to family Ruminococcaceae and the other three belong to family Lachnospiraceae. Both Ruminococcaceae and Butyricicoccus belong to order Clostridiales, which is a short-chain fatty acids (SCFA)-producing taxon. Compared with the inMPA group, the aMPA group had lower abundances of Actinobacteria, Fusobacteria and Epsilonbacteraeota at phylum level (Figure 2D, Table S2, adjusted P=0.003, 0.02, <0.001, respectively), lower abundances of Fusobacterium, Dialister, Parasutterella, Aliihoeflea, Halomonas and Bacteroides at genus level (Figure 2E, Table S3, adjusted P=0.008, <0.001, 0.038, <0.001, <0.001, <0.001, respectively) and higher abundances of Parabacteroides and Christensenellaceae at genus level (Figure 2E, Table S3, adjusted P=0.04, 0.01).
Correlation between altered genera, OTUs and clinical indices
Fourteen differential genera among the three groups were correlated with 9 clinical indices (Figure 3A). Of note, most identified taxa were corelated with serum albumin (71%), creatinine (79%) and blood urea nitrogen (64%), which partly indicated the severity of renal damage. The absolute values of the corresponding correlation coefficients were 0.33–0.49, 0.34–0.63, 0.34–0.58, respectively (Table S4). Genus Actinomymyces, Aliihoeflea, Dialister, Halomonas and Pelagibacterium were positively corelated with serum albumin and hemoglobin, and negatively corelated with serum creatinine and blood urea nitrogen. Genus Actinomyces was negatively corelated with BVAS, while Butyricicoccus was positively corelated with BVAS.
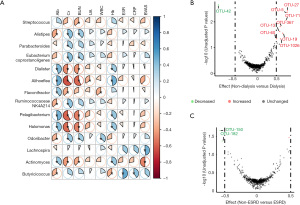
Differential analysis of OTU abundance showed that OTU-5, OTU-13, OTU-19, OTU-27, OTU-60, OTU-71, OTU-367 and OTU-1026 were significantly decreased and OTU-42 was significantly increased in patients receiving initial dialysis (n=9) by Wilcoxon test (Figure 3B). Consistently, the 5 decreased OTUs and another 2 decreased OTUs belong to family Lachnospiraceae and Prevotellaceae respectively (Table S5), both of which play an important role in SCFA production. In addition, the levels of OTU-150 and OTU-182 were increased with an absolute value of effect size ≥0.5 in the patients progressing to ESRD during follow up (Figure 3C). However, no significant difference existed when P values were adjusted by “Benjamini Hochberg” method.
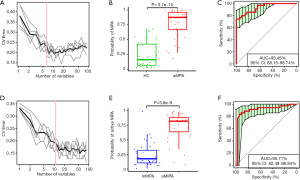
OTU markers-based models in initial diagnosis and activity evaluation of MPA
To assess the diagnostic efficiency of the gut microbiome for MPA, we incorporated OTU feature tables of the aMPA samples and the HC samples into random forest models. Five trials of 10-fold cross-validation on random forest models picked the optimal set of 6 OTU markers (Figure 4A). All these 6 OTU markers belong to Phylum Firmicutes and 4 of them belong to family Lachnospiraceae. The random forest model based on this set of OTUs verified a significantly increased predicted possibility of MPA in the aMPA samples versus the HC samples (Figure 4B, P=5.7×10–10, Wilcoxon test), and the AUC of ROC reached 93.45% (Figure 4C, 95% CI, 88.15–98.74%). We also constructed a model to distinguish the aMPA samples from the inMPA samples to evaluate disease activity with the same method. Finally, 11 OTU markers were picked (Figure 4D). This optimal set of OTUs also contributed to a model with a significantly increased predicted possibility of active disease in the aMPA samples versus the inMPA samples (Figure 4E, P=3.8×10–9, Wilcoxon test), and the AUC of ROC reached 90.71% (Figure 4F, 95% CI, 82.49–98.94%).
Discussion
MPA is a systemic inflammatory disease usually accompanied by severe renal damage (1,4,31). With advances in high-throughput sequencing, the landscape of previously uncultured microorganisms is gradually unveiled (32), and the significant role of the gut microbiome in a variety of disorders, including immune-mediated diseases (13,33,34) and CKD (15) has been illustrated. In the present study, we reported the profiling of the gut microflora in MPA patients, and further explored its clinical association and value in disease classification.
First, our results revealed that MPA patients had intestinal dysbiosis, which seemed to partially recover considering a significant rise of Shannon and Chao1 indices in inMPA versus aMPA. However, β-diversity analysis showed biased community constitution among the three groups, implying that the gut microflora of remissive MPA patients may be interfered by other cofounding factors. Actually, gut microbiota has been proven susceptible to multiple elements, such as age (35), dietary habit (36), illness (16,37), and use of antibiotics or non-antibiotic drugs (38). Chen et al. (39) found that RA patients receiving methotrexate and hydroxychloroquine had an increase in species richness and diversity. In the present study, we didn’t identify an association between use of immunosuppressants at sampling and microbial richness or diversity. It may be due to a small population, a wide variety of course and concurrent use of other drugs.
Microbial fermentation of indigestible carbohydrates can lead to the generation of SCFAs (8). Bacteria derived SCFAs have versatile functions on human physiology, such as weight control, glucose homeostasis and immunity modulation (8,37). In the present study, several SCFA-producing microbes were markedly decreased in MPA patients. Among these bacteria, Genus Roseburia, Eubacterium hallii, Anaerostipes and Butyricicoccus were implicated in the production of butyrate (8,40). Butyrate is essential for intestinal homeostasis and has immune-suppressive and anti-inflammation effects (8,37). In addition, 7 out of 8 non-dialysis enriched OTUs belonged to SCFA-producing bacteria, suggesting decreased production of SCFA may underlie the development of renal injury in MPA.
On the other hand, CKD and related alterations of commensals could impair the integrity of intestinal barrier, promote translocation of toxic compounds into circulation, and thereby induce systemic inflammation and immune paralysis (15). Wang and colleagues (16) illustrated that two ESRD-enriched species Eggerthella lenta and Fusobacterium nucleatum, could increase uremic toxins production and aggravate renal fibrosis and oxidative stress in a CKD rat model. Consistently, our data showed that genus Eggerthella was enriched at initial stage in those who progressed to ESRD later. In addition, genus Fusobacteria, Actinobacteria and Epsilonbacteraeota were increased in inMPA compared with aMPA and HCs, indicating alteration of these microbes might be owing to a chronic disease course. Besides, the abundance of phylum Proteobacteria, a major source of bacterial Lipopolysaccharide (LPS), was significantly higher in remissive MPA patients than HCs. Notably, LPS has been found elevated in CKD patients (11) and instrumental to augment glomerular damage in a murine model of anti-MPO glomerulonephritis (41). Herein, we also identified 5 new genera Actinomymyces, Aliihoeflea, Dialister, Halomonas and Pelagibacterium related to hemoglobin, two genera Flavonifractor, Odoribacter related to white blood cell and one genus Aliihoeflea related to uric acid. Since anemia, hyperuricemia and microinflammation were common complications in CKD (42,43), these taxa may act as potential indictors for progression of kidney disease in MPA.
Intriguingly, a common pattern of intestinal dysbiosis had been described in several immune mediated inflammatory diseases (IMIDs), including Crohn’s disease, ulcerative colitis, multiple sclerosis and RA (33). Specifically, increased abundance of genus Actinomyces, Eggerthella, Clostridium III, Faecalicoccus, and Streptococcus and decreased abundances of genus Gemmiger, Lachnospira, and Sporobacter were observed in all disease cohorts versus HCs. Particularly, the alterations of Actinomyce, Eggerthella, Streptococcus and Lachnospira was in line with our results. Therefore, the common pathogenic or beneficial microbes may be involved in the pathogenesis of paralleled diseases, and therapy targeting these microbes would be promising and suitable for wide application.
Though S. aureus was strongly linked to GPA from previous clinical and experimental studies (18,19), there still lacked convincing evidences for a connection between S. aureus and MPA, including this preliminary study. Recently, Gu et al. (44) identified a peptide from Actinomyces species which could induce crescentic nephritis in two murine models of anti-glomerular basement membrane (anti-GBM) disease via epitope mimicry. Actinomyces are gastrointestinal commensals which could lead to actinomycosis, a chronic granulomatous infectious disorder, when the mucosal integrity is disrupted (45). However, there was no clue of increased Actinomyces in MPA or in CKD. In the current study, genus Actinomyces were significantly increased in both aMPA and inMPA cohorts. Considering that ANCA presented in approximately 35% of patients with anti-GBM disease (46), whether Actinomyces could trigger anti-MPO immunity needs more basic research and epidemiological trials to verify. Besides, we also found an elevation of Streptococcus in both active and inactive MPA groups. Elevated intestinal Streptococcus has been described in patients with IgA nephropathy (47). Frequent mucosal carriage of Streptococcus may induce a glycosylation deficiency in IgA1, which is an important pathogenetic mechanism of IgA nephrology (48). It’s interesting that IgA class ANCA also play a pathogenetic role in ANCA associated vasculitis (49), thus it’s reasonable to speculate that Streptococcus may aggravate MPA in an IgA-dependent way. So, increased exposure of certain detrimental flora to intestinal immune system may activate specific immune response which underlies the pathogenesis of MPA.
Using gut microbiome as non-traumatic diagnostic tools has been attempted in compelling studies. Ren et al. (12) established a diagnostic model applying 30 microbial markers that validated strong diagnosis potential for early and advanced hepatocellular carcinoma. Li and colleagues (13) verified that the gut microbiome could also be used to distinguish active SLE from remissive SLE. Herein we established two models using machine-learning methods (12,30) to diagnose incipient MPA and predict activity level of MPA. The AUC reached more than 90% in both two models, indicating a good performance on classification.
Our study had several limitations. First, GPA patients were not enrolled due to the low incidence in China (5). Second, the proportion of kidney biopsy was relatively low, so the severity of renal injury couldn’t be fully assessed. Third, considering the old onset age of MPA, potential cardiovascular and metabolic diseases may interfere the gut microbial community. Four, most participants lived in one province, so whether the specific alterations in this study are applicable to other population remains to be determined.
In conclusion, our study revealed dysbiosis of the gut microbiome in MPA patients, particularly in patients with active disease. The alterations of microbial community in MPA demonstrated a combined composition of the disordered microbes verified in IMIDs and CKD, and a tendency toward loss of SCFA-producing bacteria. Furthermore, we established two random forest models based on gut microbial markers, showing the potential of diagnosing MPA and evaluating disease activity.
Acknowledgments
Funding: This study was supported by the funds from Primary Research and Development plan of Zhejiang Province (2020C03034) to FH, Zhejiang Medical and Health Science and Technology Project (2018258985) to LC.
Footnote
Reporting Checklist: The authors have completed the MDAR and STROBE reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-1315
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-1315
Peer Review File: Available at https://dx.doi.org/10.21037/atm-21-1315
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-1315). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Our research was conformed with the Declaration of Helsinki (as revised in 2013) and was approved by the ethics committee of the First Affiliated Hospital, Zhejiang University School of Medicine (approval No. 2017-694). Informed consents were available from all participants before sampling.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jennette JC, Falk RJ, Hu P, et al. Pathogenesis of antineutrophil cytoplasmic autoantibody-associated small-vessel vasculitis. Annu Rev Pathol 2013;8:139-60. [Crossref] [PubMed]
- Fujimoto S, Watts RA, Kobayashi S, et al. Comparison of the epidemiology of anti-neutrophil cytoplasmic antibody-associated vasculitis between Japan and the U.K. Rheumatology (Oxford) 2011;50:1916-20.
- Watts RA, Lane SE, Scott DG, et al. Epidemiology of vasculitis in Europe. Ann Rheum Dis 2001;60:1156-7. [Crossref] [PubMed]
- Scott DGI, Watts RA. Epidemiology and clinical features of systemic vasculitis. Clin Exp Nephrol 2013;17:607-10. [Crossref] [PubMed]
- Liu LJ, Chen M, Yu F, et al. Evaluation of a new algorithm in classification of systemic vasculitis. Rheumatology (Oxford) 2008;47:708-12. [Crossref] [PubMed]
- Jennette JC, Falk RJ. Pathogenesis of antineutrophil cytoplasmic autoantibody-mediated disease. Nat Rev Rheumatol 2014;10:463-73. [Crossref] [PubMed]
- Berti A, Cornec-Le Gall E, Cornec D, et al. Incidence, prevalence, mortality and chronic renal damage of anti-neutrophil cytoplasmic antibody-associated glomerulonephritis in a 20-year population-based cohort. Nephrol Dial Transplant 2019;34:1508-17. [Crossref] [PubMed]
- Koh A, De Vadder F, Kovatcheva-Datchary P, et al. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016;165:1332-45. [Crossref] [PubMed]
- Qin J, Li Y, Cai Z, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012;490:55-60. [Crossref] [PubMed]
- Jie Z, Xia H, Zhong SL, et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun 2017;8:845. [Crossref] [PubMed]
- McIntyre CW, Harrison LE, Eldehni MT, et al. Circulating endotoxemia: a novel factor in systemic inflammation and cardiovascular disease in chronic kidney disease. Clin J Am Soc Nephrol 2011;6:133-41. [Crossref] [PubMed]
- Ren Z, Li A, Jiang J, et al. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut 2019;68:1014-23. [Crossref] [PubMed]
- Li Y, Wang HF, Li X, et al. Disordered intestinal microbes are associated with the activity of Systemic Lupus Erythematosus. Clin Sci (Lond) 2019;133:821-38. [Crossref] [PubMed]
- Carrero JJ, González-Ortiz A, Avesani CM, et al. Plant-based diets to manage the risks and complications of chronic kidney disease. Nat Rev Nephrol 2020;16:525-42. [Crossref] [PubMed]
- Meijers B, Evenepoel P, Anders HJ. Intestinal microbiome and fitness in kidney disease. Nat Rev Nephrol 2019;15:531-45. [Crossref] [PubMed]
- Wang X, Yang S, Li S, et al. Aberrant gut microbiota alters host metabolome and impacts renal failure in humans and rodents. Gut 2020;69:2131-42. [Crossref] [PubMed]
- Chen M, Kallenberg CG. The environment, geoepidemiology and ANCA-associated vasculitides. Autoimmun Rev 2010;9:A293-8. [Crossref] [PubMed]
- Cohen Tervaert JW. Trimethoprim-sulfamethoxazole and antineutrophil cytoplasmic antibodies-associated vasculitis. Curr Opin Rheumatol 2018;30:388-94. [Crossref] [PubMed]
- Popa ER, Tervaert JW. The relation between Staphylococcus aureus and Wegener's granulomatosis: current knowledge and future directions. Intern Med 2003;42:771-80. [Crossref] [PubMed]
- Kain R, Exner M, Brandes R, et al. Molecular mimicry in pauci-immune focal necrotizing glomerulonephritis. Nat Med 2008;14:1088-96. [Crossref] [PubMed]
- Ooi JD, Jiang JH, Eggenhuizen PJ, et al. A plasmid-encoded peptide from Staphylococcus aureus induces anti-myeloperoxidase nephritogenic autoimmunity. Nat Commun 2019;10:3392. [Crossref] [PubMed]
- Pearce FA, Craven A, Merkel PA, et al. Global ethnic and geographic differences in the clinical presentations of anti-neutrophil cytoplasm antibody-associated vasculitis. Rheumatology (Oxford) 2017;56:1962-9. [Crossref] [PubMed]
- Jennette JC, Falk RJ, Bacon PA, et al. 2012 revised International Chapel Hill Consensus Conference Nomenclature of Vasculitides. Arthritis Rheum 2013;65:1-11. [Crossref] [PubMed]
- Mukhtyar C, Lee R, Brown D, et al. Modification and validation of the Birmingham Vasculitis Activity Score (version 3). Ann Rheum Dis 2009;68:1827-32. [Crossref] [PubMed]
- Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335-6. [Crossref] [PubMed]
- Edgar RC, Haas BJ, Clemente JC, et al. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011;27:2194-200. [Crossref] [PubMed]
- Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 2013;10:996-8. [Crossref] [PubMed]
- Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 2013;41:D590-6. [Crossref] [PubMed]
- Wang Q, Garrity GM, Tiedje JM, et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007;73:5261-7. [Crossref] [PubMed]
- Feng Q, Liang S, Jia H, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun 2015;6:6528. [Crossref] [PubMed]
- Watts RA, Mahr A, Mohammad AJ, et al. Classification, epidemiology and clinical subgrouping of antineutrophil cytoplasmic antibody (ANCA)-associated vasculitis. Nephrol Dial Transplant 2015;30:i14-22. [Crossref] [PubMed]
- Fraher MH, O'Toole PW, Quigley EM. Techniques used to characterize the gut microbiota: a guide for the clinician. Nat Rev Gastroenterol Hepatol 2012;9:312-22. [Crossref] [PubMed]
- Forbes JD, Chen CY, Knox NC, et al. A comparative study of the gut microbiota in immune-mediated inflammatory diseases-does a common dysbiosis exist? Microbiome 2018;6:221. [Crossref] [PubMed]
- Qian X, Liu YX, Ye X, et al. Gut microbiota in children with juvenile idiopathic arthritis: characteristics, biomarker identification, and usefulness in clinical prediction. BMC Genomics 2020;21:286. [Crossref] [PubMed]
- Bana B, Cabreiro F. The Microbiome and Aging. Annu Rev Genet 2019;53:239-61. [Crossref] [PubMed]
- Zmora N, Suez J, Elinav E. You are what you eat: diet, health and the gut microbiota. Nat Rev Gastroenterol Hepatol 2019;16:35-56. [Crossref] [PubMed]
- Kayama H, Okumura R, Takeda K. Interaction Between the Microbiota, Epithelia, and Immune Cells in the Intestine. Annu Rev Immunol 2020;38:23-48. [Crossref] [PubMed]
- Weersma RK, Zhernakova A, Fu J. Interaction between drugs and the gut microbiome. Gut 2020;69:1510-9. [Crossref] [PubMed]
- Chen J, Wright K, Davis JM, et al. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med 2016;8:43. [Crossref] [PubMed]
- Duncan SH, Barcenilla A, Stewart CS, et al. Acetate utilization and butyryl coenzyme A (CoA):acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Appl Environ Microbiol 2002;68:5186-90. [Crossref] [PubMed]
- Huugen D, Xiao H, van Esch A, et al. Aggravation of anti-myeloperoxidase antibody-induced glomerulonephritis by bacterial lipopolysaccharide: role of tumor necrosis factor-alpha. Am J Pathol 2005;167:47-58. [Crossref] [PubMed]
- Chonchol M, Shlipak MG, Katz R, et al. Relationship of uric acid with progression of kidney disease. Am J Kidney Dis 2007;50:239-47. [Crossref] [PubMed]
- Drüeke TB, Parfrey PS. Summary of the KDIGO guideline on anemia and comment: reading between the (guide)line(s). Kidney Int 2012;82:952-60. [Crossref] [PubMed]
- Gu QH, Huynh M, Shi Y, et al. Experimental Antiglomerular Basement Membrane GN Induced by a Peptide from Actinomyces. J Am Soc Nephrol 2020;31:1282-95. [Crossref] [PubMed]
- Könönen E, Wade WG. Actinomyces and related organisms in human infections. Clin Microbiol Rev 2015;28:419-42. [Crossref] [PubMed]
- DE Zoysa J. Incidence and features of dual anti-GBM-positive and ANCA-positive patients. Nephrology (Carlton) 2011;16:725-9. [Crossref] [PubMed]
- Wu H, Tang D, Zheng F, et al. Identification of a novel interplay between intestinal bacteria and metabolites in Chinese patients with IgA nephropathy via integrated microbiome and metabolome approaches. Ann Transl Med 2021;9:32. [Crossref] [PubMed]
- Naka S, Wato K, Misaki T, et al. Streptococcus mutans induces IgA nephropathy-like glomerulonephritis in rats with severe dental caries. Sci Rep 2021;11:5784. [Crossref] [PubMed]
- Kelley JM, Monach PA, Ji C, et al. IgA and IgG antineutrophil cytoplasmic antibody engagement of Fc receptor genetic variants influences granulomatosis with polyangiitis. Proc Natl Acad Sci U S A 2011;108:20736-41. [Crossref] [PubMed]


