
Development and validation of Auto-Neo-electroencephalography (EEG) to estimate brain age and predict report conclusion for electroencephalography monitoring data in neonatal intensive care units
Introduction
In neonatal intensive care units (NICUs), continuous electroencephalography (EEG) monitoring has been widely applied to the diagnosis of neonatal neurological diseases such as epilepsy, encephalopathy, and central nervous system infection (1,2). Also, the long-term neurological outcome is associated with early-onset EEG changes (3). Though it is easy to perform EEG monitors on neonates lasting for hours or even days at the bedside, the raw EEG signal data are usually very large, which takes experienced neurophysiologists several hours to interpret. Besides, that neonates’ brain is developing complicates the evaluation of EEG pattern (4), especially among preterm neonates (5). O’Reilly designed a new EEG signal feature range EEG (rEEG) in 26 newborns with less than 29 weeks of gestational age and found that it is closely related to brain development and maturity (6). Similarly, Stevenson et al. constructed a brain age prediction model based on EEG signal features from 65 preterm infants, which could greatly fit actual age and the predicted age difference could be used as a predictor of the neurodevelopmental outcome (5,7,8). These studies are all tested on small data sets, and there is no systematic analysis on how to apply the findings to the clinic.
Developing a machine learning strategy that can quantitatively analyze the EEG signal dataset is crucial, which could make automatically screening the abnormal possible to assist clinicians in EEG report generation and further diagnosis. Therefore, in this study, we aimed to construct an automatic system that could uncover brain age and suggest abnormality from original EEG signals. Here, we collected a large group of EEG datasets from the NICU at Children’s Hospital of Fudan University, including 1,851 subjects with signal data and clinical reports. We constructed a system named Auto-Neo-EEG, which consists of an EEG neural signal processing pipeline to extract features from the original signal datasets, machine learning models based on gradient boosted model (gbm) (9) for prediction. The model could achieve great performance on estimating brain age for neonates and figuring out abnormal EEG records, showing great potential in NICU application.
We present the following article in accordance with the STARD reporting checklist (available at https://dx.doi.org/10.21037/atm-21-1564).
Methods
Patients
We retrospectively reviewed patients who underwent EEG monitoring in the NICU in Children’s Hospital of Fudan University, from Jan. 2016 to Mar. 2018. A total of 1,851 subjects from 1,692 patients were collected. The study was approved by the ethics committee of the Children’s Hospital of Fudan University (No. 2020227), the patients recruited belong to a neonatal project (NCT02544100), and written informed consent was obtained from the guardians of each patient. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Inclusion and exclusion criteria
Shown in Figure 1A, the inclusion criteria for subjects were as follows: (I) neonates whose conceptional age (CA) ranged from 29+0 to 44+6 weeks (203 to 314 days) at the beginning of the recording period; (II) neonates who had video-continuous EEG examinations with a valid observation span over 30 minutes; (III) the first recording was taken if a patient had several recordings within one day. The exclusion criteria were: (I) neonates whose EEG clinical reports were missing or incomplete in some necessary signals.
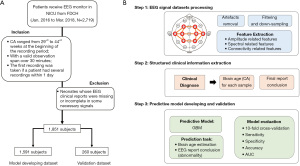
Dataset acquisition
EEG data were acquired using a Nicolet One machine (sampling frequency: 500 Hz). We followed the International 10–20 system to place the electrodes. The frontal (F3, F4), central (C3, C4), mid-temporal (T3, T4), parietal (P3, P4) scalp electrodes and reference electrode (Cz) was placed (10). We chose the parietal area (P3/P4) instead of the occipital area (O1/O2) because more artifacts detected in the occipital area.
We had three experienced clinicians (Y Zhou, X Wang, Y Xu) in charge of the EEG report generation, who all had attended the uniformly training program and were certified by the Chinese Anti-Epilepsy Association. Y Zhou is the senior clinician with more than 10 years of experience in neonatal EEG reading, and X Wang and Y Xu both have 5 years of experience. For each EEG clinical interpretation, X Wang and Y Xu would perform a double-blind interpretation of the EEG signal and make a conclusion. If the result from the two experts (X Wang and Y Xu) was consistent, they would directly apply it as the final report. If not, Y Zhou will join and they three would discuss it together to make a final decision.
Standard-scheme for manually reported EEG conclusion
The clinicians followed the guideline—a handbook for clinical EEG reading (11)—to give a clinical report. The levels of EEG abnormalities are defined as slightly abnormal, moderately abnormal, and severely abnormal, and the details are as follows:
Slightly abnormal
- Compared with the actual CA, the background activity is mature but slightly delayed, and the trace alternant (TA) or Tracé discontinu (TD) are slightly discontinuous;
- The waveform or rhythm compatible with CA is slightly lacking, or the immature waveform compatible with CA disappears;
- Focal electrical attenuation;
- A small number of focal or multifocal discharges on a normal or mildly abnormal background.
Moderately abnormal
- Compared with the actual CA, the background activity is moderately discontinuous [for neonates whose CA is less than 30 weeks, the interval between bursts is over 30 s; for neonates over 30 weeks (CA), the interval is between 20 and 60 s];
- The waveform or rhythm compatible with CA is lacking, or the immature waveform compatible with CA obviously disappears;
- The hemisphere continues to be asymmetrical and/or out of sync, but not exceeding 50% of the entire record;
- Continuous universal voltage reduction, the background activity below 25 µV in all states;
- Single-disciplinary or other forms of electrical attack without severe background abnormalities;
- Other abnormality whose level is between slightly and moderately abnormal but does not belong to the above descriptions of moderately abnormal, such as multiple discharge waveform.
Severely abnormal
- Compared with actual CA, the background activity is significantly discontinuous (for example, the burst interval exceeds 60 s);
- Focal or one-sided periodic discharge exists;
- Over-synchronization and or asymmetry between hemispheres, making up more than 50% of the entire record;
- Sharp waves frequently appear in the Rolandic or midline area;
- Severely low voltage (below 5 µV in all states);
- Burst suppression;
- Equipotential.
Interrater agreement assessment
Here, we randomly selected 96 subjects for detailed interrater agreement assessment by two experts (X Wang, Y Xu). Cohen’s Kappa was used to investigate interrater agreement level for binary level and weighted Kappa was used for ordinal report level. The test was realized by using Kappa function from R package vcd (version 1.4-7).
Establishment of Auto-Neo-EEG pipeline
In total, 1,851 subjects were recruited for analysis, of which 1,591 subjects were used in a retrospective study for prediction model generation, while the remaining 260 were included in the validation dataset. The model-developing dataset was used to generate models, including the optimization of model hyper-parameters, while the validation dataset, independent from the model training procedure, was used to evaluate prediction performance.
As shown in Figure 1B, the Auto-Neo-EEG pipeline consists of:
EEG signal pre-processing and feature extraction from EEG dataset
For each EEG recording, the original signal dataset was pre-processed, including adjusted to the reference electrode, artifact removal, filtering, and down-sampling. The artifact removal steps were as follows: (I) improper electrode placement: Improper electrode placement can lead to the removal of channels with low correlation coefficients with all the other channels (the threshold was set at 0.15). (II) Electrode coupling: electrode coupling mainly allowed the identification of channels with relatively low power compared with all channels from the same hemisphere. (III) Continuous zeros or constant values: continuous zeros or constant values may result from procedures such as testing the electrode impedance, and these values should be removed. (IV) High amplitude & (V) sudden jump removal: the high amplitude and sudden jump removal steps mainly focused on segments with abnormally high values or value changes possibly caused by movement. The thresholds were 1,500 µV for the absolute amplitude and 200 µV for the amplitude difference, which are also suitable for preterm infants. In the first two steps, a whole channel was removed, and the values of the other channels changed to “NA” (not available) for the segments that failed the filtering procedure. (VI) Too many “NA” values: all “NA” values across all channels were examined and segments with an excessive number of “NA” values were removed. (VII) Improper correlations between independent component analysis (ICA) and electrooculography (EOG): the last step was performed if the original dataset contained an EOG channel. ICA was used to decompose the EEG signal into independent components, and each component was compared to the EOG channel to identify any improper correlations. The effects of the rejected components were removed from the original data. The first five steps were reported by O’Toole et al. (12).
Then, a notch filter was performed at 50, 100, 150, 200, and 250 Hz; and a finite-impulse response (FIR) filter was applied in a range between 0.3 to 50 Hz. Finally, the dataset was down-sampled from 500 to 100 Hz.
A total of 722 signal features were extracted to reflect the amplitude, rEEG, spectral density, and connectivity-related aspects. Firstly, the original signal dataset could be decomposed into four frequency bands, i.e., 0.5–4, 4–8, 8–13, and 13–30 Hz, and the number of signal channels was eight. For each frequency band and in each channel, six amplitude features (amplitude_total_power, amplitude_SD, amplitude_skew, amplitude_kurtosis, amplitude_env_mean, amplitude_env_sd), eight rEEG features (rEEG_mean, rEEG_median, rEEG_lower_margin, rEEG_upper_margin, rEEG_width, rEEG_SD, rEEG_CV, rEEG_asymmetry), five spectral features (spectral_power, spectral_relative_power, spectral_flatness, spectral_entropy, spectral_diff) were extracted, thus the first number of signal features was 608 [(6+8+5)*4 (frequency band)*8 (channel)]. Secondly, two spectral related features (spectral_edge_frequency, FD) were extracted in each of eight channels: 2*8=16. For the above aspects, the average figure for all channels was also calculated as new features, which resulted in (6+8+5)*4+2=78 more features. Besides, five connectivity related features (connectivity_BSI, connectivity_corr, connectivity_coh_mean, connectivity_coh_max, connectivitiy_coh_freqmax) were extracted for each of four frequency bands (5*4=20). In total, 722 (608+16+78+20) neural signalling features were obtained for each sample.
Figure S1 shows the flowchart of the signal processing procedure. We referred to the research by O’Toole et al. (12) to perform some of the signal processing steps. The processing steps were developed based on the Python3.6 environment with MNE (13).
EEG signal finding extraction from clinical reports
In clinical reports, CA and report conclusions were extracted. CA was normalized to days and EEG report conclusions were labelled as one of the four ordinal categories (normal, slightly abnormal, moderately abnormal, and severely abnormal).
Prediction model outcome declaration and evaluation criteria
We had two tasks: CA estimation and report conclusion (abnormal severity) prediction, and the outcomes were declared below:
CA estimation; the observed CA was normalized into days and the machine learning model could directly output the predicted days. We built the prediction model merely using the samples with a normal conclusion from the model-developing dataset. The comparison between the predicted CA and the observed CA would be measured by Pearson correlation coefficient (PCC) with significance and 95% confidence interval (CI).
Report conclusion (abnormal severity) prediction; since it was a multi-class prediction issue, we adopted the cascade strategy, i.e., we transformed the classes into four bi-classification questions based on abnormal severity: severely abnormal vs. the rest, moderately abnormal vs. normal/slightly abnormal, slightly abnormal vs. normal, and abnormal vs. normal. The final predicted result would be severely abnormal if the first strategy (severely abnormal vs. the rest) was severely abnormal. The final predicted result would be moderately abnormal if the first strategy (severely abnormal vs. the rest) was the rest and the second strategy (moderately abnormal vs. normal/slightly abnormal) was moderately abnormal. The final predicted result would be slightly abnormal if the first two strategies were the rest and normal/slightly abnormal but the third strategy (slightly abnormal vs. normal) was slightly abnormal. If the result from the third strategy was normal, the sample would be classified as normal. Combining the above three binary classification models, we could give a unique final label for each sample. The fourth strategy was set as a reference for clinician judgment (if the prediction system user does not want to classify the abnormality with three different levels), and this would not be used in the report conclusion judgment in our study. We calculated area under the curve (AUC) under receiver operating characteristic (ROC) curve with 95% CI, sensitivity, specificity, and accuracy in the model-developing dataset and an independent validation dataset to evaluate the model’s performance.
Machine learning steps
Dataset splitting
We split the dataset according to the sample collection time. 1,591 subjects before 2018 were used as the model-developing dataset and the remaining 260 subjects after 2018 were treated as the independent validation dataset.
Machine learning model selection and results evaluation
The classification and regression prediction tasks were all based on the gbm. As described above, PCC was used to evaluate the performance of regression prediction tasks and AUC under ROC curve with sensitivity, specificity, and accuracy at the optimal threshold was used to evaluate the performance of binary classification tasks. For the report-conclusion prediction with four levels, the confusion matrix was generated with sensitivity, specificity, and accuracy to evaluate the performance.
Feature selection
A backward selection procedure was applied. For each iteration, gbm model was generated and features with the minimum importance value were removed. The remained features from the model with the best performance for the cross-validation (CV) results in the model-developing dataset were used. The feature selection was performed separately for different prediction tasks.
Model generation and CV
In the model-developing dataset, we applied a 10-fold CV strategy for training the prediction model. All predictions were realized by gbm and within each CV, different value combinations for each of the parameters would be iterated, i.e., interaction.depth, n.trees, shrinkage, and n.minobsinnode. The final model was the one with the best performance (highest PCC value or AUC value) for the CV results.
For the final used model, there would be “importance values” to show the signal feature importance with a higher value to indicate more contribution in the prediction model. For binary classification, the “Youden’s J statistic” was employed to get the optimal cut-off. The selected final model with the optimal threshold was applied in both the model-developing and independent validation dataset to estimate the performance.
Code implementation
All scripts for prediction and visualization were written in R version 3.6, with packages caret (https://cran.r-project.org/package=caret) and gbm (https://cran.r-project.org/package=gbm).
Statistical analysis
We applied a two-tailed Student’s t-test for comparison of continuous variables between two groups and an F-test for overall significance in linear regression. Fisher’s Exact test was used for the enrichment testing for 2*2 categorical data. Statistical significance was defined as P<0.05 and false discovery rate (FDR) correction was used for multiple tests. Statistical test in the model generation procedure is described above. All statistical analyses were performed using R version 3.6.
Results
Benchmark EEG dataset
A total of 1,851 subjects of video-continuous EEG recordings were recruited (Figure 1A), with the corresponding clinical reports and disease diagnoses collected. The basic statistics were summarized in Table 1. The detailed clinical reports about 30 recording findings could result in a four-level report conclusion judged by experienced clinicians (Table S1 and Method section). The number of samples with four categories of EEG report conclusions in each corrected gestational age (CA) week was shown in Figure S2. The average monitoring time was 2.7 hours. The corresponding 1,692 patients (some patients had multiple recordings) were finally diagnosed (with the help of EEG recordings and other clinical diagnoses) and classified into 11 disease systems. Hypoxic ischemic encephalopathy (HIE), central nervous system infection, congenital metabolic disease and unexplained convulsions tend to have moderate and severely abnormal EEG report conclusion, while temporary metabolic disorder tends to be normal (Table S2, P value <0.05). In the following sections, we would build an intelligent system—Auto-Neo-EEG (the detailed process is shown in Figure 1B and Figure S1)—to systematically investigate the correlation between EEG signals and CA in neonates, and based on that we generate a predictive model to facilitate EEG report generation.
Full table
For the Interrater agreement assessment, 96 randomly selected subjects were shown in Table S3. Only nine subjects were inconsistent for report conclusion level between two experts (patients 5, 8, 48, 58, 63, 66, 92, 93, 96) with 90.63% agreement percentage. The weighted Kappa value was 0.923 if consider the ordinal report level and the Cohen’s Kappa value was 0.913 if merge three abnormal levels into one. Generally, the good interrater agreement level for outcome ensures the quality of the benchmark dataset used in this study.
Auto-Neo-EEG could successfully estimate brain age
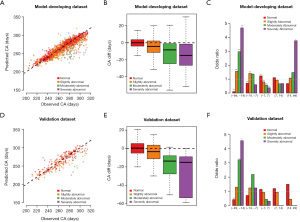
In total, 59 signal features passed the signal filtering step were used to fit the prediction model which achieved high accordance with PCC 0.966 (95% CI: 0.961–0.970, P value <2e−16, F-test) in identifying the CA for 845 datasets with normal reported conclusion level (red points in Figure 2A), but the difference occurs for samples with not-normal conclusion levels (orange, green and purple points in Figure 2A). When estimating the deviation—CA_diff (difference between the predicted and real CAs), it was significantly less than zero in all three abnormal groups, and the differences increased with the severity of the abnormality (Figure 2B, all P value <1e−6, Student’s t-test, with median values −4.27, −8.36, and −14.90). The severely abnormal samples were significantly enriched in groups with CA difference smaller than 14 days, followed by moderately abnormal (Figure 2C, P value <0.05). Samples with CA difference larger than 14 days were also enriched in severely abnormal (Figure 2C, P value <0.05). The model also achieved great performance in the validation dataset with accordance of PCC 0.904 (95% CI: 0.870–0.930, P value <2e−16, F-test, red points in Figure 2D) for the samples with normal conclusion level. Similarly, the CA_diff decreases with the severity of abnormality (Figure 2E, all P value <0.05, Student’s t-test, with median values −3.46, −13.71, and −15.12). Further, we divided all samples into five groups according to the difference between the predicted CA and actual CA {(−Inf, −14], (−14, −7], (−7,7], (7,14], (14, Inf]} and calculated the odds ratio for each EEG report level with statistical testing. Similarly, in the validation dataset, the severely abnormal samples were significantly enriched in groups with CA difference smaller than 14 days, followed by moderately abnormal (Figure 2F, P value <0.05). We tried to directly apply the absolute CA difference as the predictor to the four EEG reporting levels by using “Youden’s J statistic” to get the optimal threshold, the result showed that the absolute CA difference was a good marker for the severely abnormal prediction (Table S4, accuracy >80%). Therefore, we could apply Auto-Neo-EEG to quantitative estimation for EEG maturity, and the deviation to the real CA highlights the severity of the abnormality.
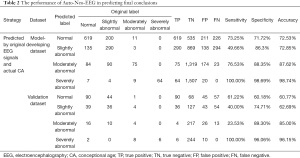
Auto-Neo-EEG could achieve high performance to classify EEG abnormality
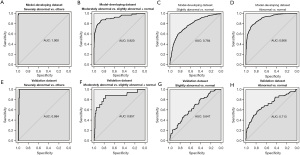
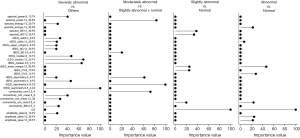
Next, we developed models to predict four levels of EEG report conclusions directly from the original EEG signals. As we applied the cascade strategy with four binary comparisons to deal with the multi-class classification issues, the original pairwise performance was shown in Figure 3 and Table S5 with the final confusion matrix shown in Table 2. The best prediction accuracy was achieved in the severely abnormal category (AUC 1 for the model-developing dataset with 95% CI: 0.999–1.000, 0.984 the validation dataset with 95% CI: 0.970–0.999), followed by the moderately abnormal category (AUC 0.919 for the model-developing dataset with 95% CI: 0.885–0.955 and 0.857 for the validation dataset with 95% CI: 0.741–0.973). Distinguishing of these two levels could achieve great accuracy and specificity (all higher than 85%), but relatively low sensitivity was observed in moderately abnormal, where several samples were over-estimated to the severely abnormal level. Differentiating the normal and slightly abnormal conditions tended to be more challenging than that in other conditions, which may be due to slight abnormalities in clinical judgments that are relatively more subjective (Figure 3C,3G, Table S5). Besides, the prediction model separating abnormal (combining three abnormal levels) and normal subjects only achieved ordinary performance (Figure 3D,3H, Table S5). The features that contributed the most to identify the severely abnormal conditions (with highest importance value) were rEEG (similar to aEEG) lower median and asymmetry, whereas to identifying the slightly abnormal conditions, the observed CA and spectral difference contributed the most (Figure 4). Generally, Auto-Neo-EEG could successfully predict EEG report conclusion abnormality.
Full table
Discussion
Continuous EEG as a non-invasive tool of brain function monitoring in the NICU is recommended by the American Clinical Neurophysiology Society (ACNS) (14,15). In acute neonatal encephalopathy, EEG can provide useful information on brain function. Severe background activity abnormalities reflect serious brain damage and are prognostic predictors of long-term adverse outcomes (1). Awal et al reviewed 52 studies and showed that burst suppression and low voltage could accurately predict the neurologic sequelae of newborns with HIE (16). Moreover, few biomarkers have been used to assess brain maturation in neonates (17). To data, magnetic resonance imaging (MRI) is the only clinical tool for assessing the brain integrity in neonates (18). Recently, one study showed that preterm EEG can be used to estimate functional brain age. Therefore, continuous EEG monitoring is critical in NICU. However, many difficulties still exist in the clinics. For example, neonatal EEG interpretation requires a specialized neurophysiologist who, however, definitely not be available to interpret EEG signals 24/7 (19). To address the gap in EEG monitoring and interpretation, real-time and automatic equipment to interpret EEG data is needed.
Here, the Auto-Neo-EEG, which focuses on automatic brain age estimation and classifying the EEG abnormality greatly accelerates the generation of EEG reports for neonates. Our findings suggested that Auto-Neo-EEG could predict brain maturity and classify the EEG abnormality with high accuracy. This system has multiple advantages. First, it is generated according to a large-scale dataset. Children’s Hospital of Fudan University has launched EEG monitoring in NICU for 7 years and has assisted over 5,000 patients in disease diagnosis so far. We have six Nicolet One machines with the specially assigned technologist to perform EEG signal recording, which has guaranteed the effectiveness in the data collection procedure. We found clear correlation patterns between CA and signal patterns and uncovered the relationship between CA difference and abnormal EEG signals quantitatively, which could directly be applied to automatic maturity abnormality screening. We also tried to directly apply the CA difference to predict the EEG report conclusion (Table S4), which showed that the performance to distinguish severely abnormal and moderately abnormal was much better than normal and slightly abnormal but still worse than the results shown in Table 2, indicating that the difference for CA was a good marker for the abnormality prediction but not the only affected signal features. Besides, it has been previously reported that EEG interpretation lacks consistency (20), and in our dataset, the interrater agreement for the report conclusion could reach a perfect level.
Motivated by clinical requirements, we integrated the diagnostic information of patients, clinical reports, and original EEG signal datasets to build a prediction system to assist in generating neonatal EEG reports. We have made considerable progress in the following aspects: (I) we have established a standardized platform for data collection and structured clinical report generation. (II) The Auto-Neo-EEG is an automated, objective, 24/7, and standardized interpretation of bedside EEG monitors, which could effectively assist in clinical report generation. (III) The model to identify CA could provide a quantitative estimation of brain age, which could hardly be judged using traditional strategy. The difference between predicted and observed CA could strongly indicate abnormality as lots of factors such as asphyxia, multi-system malformation will delay brain development. This model could directly be applied without much manual review. In addition, the choice of predictive models: we chose a gradient boosting machine (gbm) as the predictive model in our study and the EEG report conclusion prediction task convert the original multi-class classification issue into three binary classification issues. We also tried some popular algorithms, such as lasso regression for CA prediction and random forest and support vector machine (SVM) for bi-classification prediction, and the performance of those algorithms were worse than that of gbm. We also tried to generate a model to directly predict the four report levels, but the results were worse than the current model, especially in the prediction of severely and moderately abnormal. In addition, we tried deep learning frameworks EEGLearn (21), which had a similar performance, but additional computing resources were required.
Limitations
One of the major limitations in our system is the criteria used in the clinical report generation to train the prediction model, rendering it could only generate reports that follow Liu’s guideline, which is generally followed the description of table 6.3 from Ebersole et al. (8). Besides, some technical limitations are as follows. (I) Local conclusion prediction: we tried to locate abnormal signal features in specific periods, but the result was not ideal (the false positive rate is high), partially due to lack of accurate label, and more diverse data collections and accurate manual annotations are required in future improvement. (I) Signal finding prediction: we are trying to learn from prediction algorithms, such as an empirical wavelet transformation applied by Bhattacharyya et al. (22), sharp and wave calculations applied by Chang et al. (23), different types of entropies mentioned by Arunkumar et al. (24), and key-point based local binary patterns proposed by Tiwari et al. (25), to further broaden the signal description prediction in the clinical report. (III) Impact of medications: we do not consider the potential impact of medications on the EEG signals in this study, which we will systematically design and discuss the effect in our future work. (V) Clinical application: currently our system still needs clinical experts to review before a clinical report is done. During this process, the original signal data visualization and a report review system for clinical manipulation are required. Thus, we are developing a server that will include the EEG signal browser, a management system for patient information, and a report review system. The predictive model can also be updated with more data collected during its further clinical application.
Conclusions
In conclusion, the present study shows that Auto-Neo-EEG can successfully estimate brain age and predict signal abnormalities, which could benefit many clinicians in performing neonatal EEG studies.
Acknowledgments
We thank all doctors and nurses in NICU for their patient care and data collection.
Funding: This work was funded by the project supported by Shanghai Municipal Science and Technology Major Project (2018SHZDZX05) and National Key Research and Development Project of China (2018YFC0116903). These two funding had helped in the data collection procedure by paying the nurses who operated the machine and helped the data analysis procedure in purchasing the computer clusters for large-scale data storage and computing. Intelligent Medical Research Project of Shanghai Health and Family Planning Commission (2018ZHYL0225). This funding has helped in the data collection procedure by paying the clinician and researchers to collect and clean the original EEG dataset and clinical report, to perform the clinical report decomposing and label revision.
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-1564
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-1564
Peer Review File: Available at https://dx.doi.org/10.21037/atm-21-1564
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-1564). Xinran Dong and Yulan Lu report that funding (2018YFC0116903 and 2018ZHYL0225) was received to support this manuscript. Yanting Kong and Guoqiang Cheng report funding (2018ZHYL0225) was received to support this manuscript. Wenhao Zhou report funding (2018SHZDZX05 and 2018YFC0116903) was received to support this manuscript. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of the Children’s Hospital of Fudan University (No. 2020227), the patients recruited belong to a neonatal project (NCT02544100), and written informed consent was obtained from the guardians of each patient.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Massey SL, Jensen FE, Abend NS. Electroencephalographic monitoring for seizure identification and prognosis in term neonates. Semin Fetal Neonatal Med 2018;23:168-74. [Crossref] [PubMed]
- McCoy B, Hahn CD. Continuous EEG monitoring in the neonatal intensive care unit. J Clin Neurophysiol 2013;30:106-14. [Crossref] [PubMed]
- Périvier M, Rozé JC, Gascoin G, et al. Neonatal EEG and neurodevelopmental outcome in preterm infants born before 32 weeks. Arch Dis Child Fetal Neonatal Ed 2016;101:F253-9. [Crossref] [PubMed]
- Mathieson SR, Stevenson NJ, Low E, et al. Validation of an automated seizure detection algorithm for term neonates. Clin Neurophysiol 2016;127:156-68. [Crossref] [PubMed]
- Stevenson NJ, Oberdorfer L, Tataranno ML, et al. Automated cot-side tracking of functional brain age in preterm infants. Ann Clin Transl Neurol 2020;7:891-902. [Crossref] [PubMed]
- O'Reilly D, Navakatikyan MA, Filip M, et al. Peak-to-peak amplitude in neonatal brain monitoring of premature infants. Clin Neurophysiol 2012;123:2139-53. [Crossref] [PubMed]
- Stevenson NJ, Oberdorfer L, Koolen N, et al. Functional maturation in preterm infants measured by serial recording of cortical activity. Sci Rep 2017;7:12969. [Crossref] [PubMed]
- Ebersole JS, Pedley TA. Current practice of clinical electroencephalography. 3rd edition. Philadelphia: Lippincott Williams & Wilkins, 2003.
- Friedman JH. Greedy function approximation: A gradient boosting machine. Ann Statist 2001;29:1189-232. [Crossref]
- Homan RW, Herman J, Purdy P. Cerebral location of international 10-20 system electrode placement. Electroencephalogr Clin Neurophysiol 1987;66:376-82. [Crossref] [PubMed]
- Liu X. Clinical Electroencephalography (version II). Beijing: People’s Medical Publishing House, 2019.
-
O'Toole JM Boylan GB NEURAL: quantitative features for newborn EEG using Matlab. arXiv:1704.05694[physics.med-ph] 2017 . - Gramfort A, Luessi M, Larson E, et al. MNE software for processing MEG and EEG data. Neuroimage 2014;86:446-60. [Crossref] [PubMed]
- Shellhaas RA, Chang T, Tsuchida T, et al. The American Clinical Neurophysiology Society's Guideline on Continuous Electroencephalography Monitoring in Neonates. J Clin Neurophysiol 2011;28:611-7. [Crossref] [PubMed]
- Nagarajan L, Palumbo L, Ghosh S. Classification of clinical semiology in epileptic seizures in neonates. Eur J Paediatr Neurol 2012;16:118-25. [Crossref] [PubMed]
- Awal MA, Lai MM, Azemi G, et al. EEG background features that predict outcome in term neonates with hypoxic ischaemic encephalopathy: A structured review. Clin Neurophysiol 2016;127:285-96. [Crossref] [PubMed]
- Somerville LH. Searching for Signatures of Brain Maturity: What Are We Searching For? Neuron 2016;92:1164-7. [Crossref] [PubMed]
- Pittet MP, Vasung L, Huppi PS, et al. Newborns and preterm infants at term equivalent age: A semi-quantitative assessment of cerebral maturity. Neuroimage Clin 2019;24:102014 [Crossref] [PubMed]
- Ntonfo GM, Ferrari G, Raheli R, et al. Low-complexity image processing for real-time detection of neonatal clonic seizures. IEEE Trans Inf Technol Biomed 2012;16:375-82. [Crossref] [PubMed]
- Wusthoff CJ, Sullivan J, Glass HC, et al. Interrater agreement in the interpretation of neonatal electroencephalography in hypoxic-ischemic encephalopathy. Epilepsia 2017;58:429-35. [Crossref] [PubMed]
- Bashivan P, Rish I, Yeasin M, et al. Learning Representations from EEG with Deep Recurrent-Convolutional Neural Networks. 2015;
- Bhattacharyya A, Pachori RB. A Multivariate Approach for Patient-Specific EEG Seizure Detection Using Empirical Wavelet Transform. IEEE Trans Biomed Eng 2017;64:2003-15. [Crossref] [PubMed]
- Chang WD, Cha HS, Lee C, et al. Automatic Identification of Interictal Epileptiform Discharges in Secondary Generalized Epilepsy. Comput Math Methods Med 2016;2016:8701973 [Crossref] [PubMed]
- Arunkumar N, Ramkumar K, Venkatraman V, et al. Classification of focal and non focal EEG using entropies. Pattern Recognit Lett 2017;94:112-7. [Crossref]
- Tiwari AK, Pachori RB, Kanhangad V, et al. Automated Diagnosis of Epilepsy Using Key-Point-Based Local Binary Pattern of EEG Signals. IEEE J Biomed Health Inform 2017;21:888-96. [Crossref] [PubMed]


