
miRNA-218/FANCI is associated with metastasis and poor prognosis in lung adenocarcinoma: a bioinformatics analysis
Introduction
At present, lung cancer has the highest mortality rate worldwide, and lung adenocarcinoma has become the most common form of lung cancer (1). LUAD is typically diagnosed in advanced stages, when the cancer has metastasized to nearby tissues (2). Late diagnosis leads to delayed treatment, and failure of prognosis evaluation leads to a low survival rate among LUAD patients (3). Furthermore, drug resistance has also limited the clinical use of targeted drugs. Therefore, exploring the mechanisms of LUAD progression is urgently needed, which could aid in the diagnosis and discovery of prognostic biomarkers and potential therapeutic targets (4). Numerous biomarkers and therapeutic targets have been shown to play an effective role in the mechanisms of treatment for LUAD patients. However, there remains a pressing need to identify more genetic information of the key genes in LUAD progression.
Currently, comprehensive bioinformatics analysis, which is mainly focused on screening and further analyzing differentially expressed molecules, has been widely used in the diagnosis and treatment of LUAD, as well as the identification of potential biomarkers associated with prognosis. Accumulating data on malignancy is available from the authoritative GEO genomics data repository, which includes microarray and high-throughput sequencing data. In the current study, we used one microRNA (miRNA) microarray and three messenger RNA (mRNA) microarrays to obtain DEMs and DEGs in order to reduce false positive results. Functional enrichment of DEGs and DEMs was carried out. The protein-protein interaction (PPI) network analysis was used to screen out the hub genes related to LUAD through the Molecular Complex Detection (MCODE) module. Key miRNAs and mRNAs in LUAD were identified by miRNA-mRNA interaction analysis and OS analysis. Mir-218 was been shown to be downregulated in a variety of malignant tumors, thereby playing an important function similar to that of tumor suppressor genes. In the current study, we confirmed that miR-218 is downregulated and related to poor prognosis in LUAD patients. In the hub DEGs, FANCI was observed to be an important potential prognostic biomarker for LUAD. We identified that miR-218 is negatively correlated with the FANCI expression in LUAD for the first time.
Additionally, we verified FANCI expression in LUAD using The Cancer Genome Atlas (TCGA) and GEO databases, and explored the correlation between FANCI expression and the clinicopathological characteristics as well as the prognosis of LUAD patients. Our results provide more evidence to confirm that FANCI is a LUAD biomarker that may be involved in the mechanisms of metastasis as well as the poor prognosis of LUAD patients. This may help to provide targeted therapy for LUAD patients with novel targets.
We present the following article in accordance with the REMARK reporting checklist (available at https://dx.doi.org/10.21037/atm-21-3823).
Methods
Microarray data
The GEO Series (GSE)63805 miRNA microarray and the GSE40791, GSE27262, and GSE75307 mRNA microarrays were downloaded from the GEO database. The GSE40791 dataset included 100 normal tissues and 94 LUAD tissues. GSE27262 comprised 50 samples, including 25 LUAD tissues and 25 normal tissues. GSE75037 contained 166 samples, including 83 LUAD samples and 83 normal samples. GSE63805 included 30 LUAD samples and 30 normal samples.
Clinical specimen collection
Thirty surgical specimens from LUAD patients, including lung cancer tissues and paracancerous tissues, were collected at the Department of Thoracic Surgery in the First Affiliated Hospital of Zhengzhou University from September 2019 to January 2020. These patients had primary tumors and did not receive preoperative chemoradiotherapy, biotherapy, or antitumor therapy. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University (NO.2019-KY-255), and the patients and their families agreed to the tissue samples collection and their use in research.
DEGs and DEMs analysis
In this study, GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/), a comprehensive tool that can analyze different expression data groups, was employed to identify DEGs and DEMs. An adjusted P value <0.01 and logFC >1.5 was applied for DEGs, and an adjusted P value <0.05 and log FC >1 was used for DEMs. The overlapping DEGs were derived from the GSE40791, GSE27262, and GSE75037 datasets.
Pathway analysis for DEMs and DEGs
Database for Annotation, Visualization, and Integrated Discovery (DAVID, https://david.ncifcrf.gov/), an online bioinformatics database, was used for pathway analysis of DEGs and DEMs. We then visualized the results using RStudio software (Boston, RStudio, Inc.). Similarly, enrichment analysis was performed for DEMs using FunRich (http://www.funrich.org).
Construction of the PPI network
Search Tool for the Retrieval of Interacting Genes (STRING, https://string-db.org/) is an online website that was designed to predict PPI network. In this study, we set an interaction score >0.4, and analyzed the PPI network for DEGs. We subsequently visualized the results using Cytoscape software. MCODE, a Cytoscape plugin, was used to locate the hub genes of the PPI network.
Kaplan-Meier (KM) analysis of DEMs
We used the KM plotter to perform an overall survival (OS) analysis of the DEMs in 513 and 866 LUAD patients, respectively. We separated patients into two groups according to the gene transcriptional expression level, and drew KM plots.
MiRNA target gene prediction
MiRecords (http://c1.accurascience.com/miRecords/) was used to predict the target mRNA of survivable DEMs, which consisted of 11 miRNA target gene prediction algorithms. We accepted target mRNA of DEMs that were predicted in at least three databases. Next, we visualized the regulatory network using Cytoscape (Institute for Systems Biology).
miRNA-mRNA network construction
Previous studies have demonstrated that miRNA regulated the expression of mRNA by translation inhibition or degradation, which indicated that miRNA was negatively related to miRNA expression. The reverse trend of DEM-DEG (upregulated DEM-downregulated DEG or downregulated DEM-upregulated DEG) was screened, and Cytoscape was used to visualize DEM-DEG network was used.
Relationship between FANCI and miR-218 expression
The online prediction website TargetScan (http://www.targetscan.org/vert_72/) was used to predict the miRNA target genes and verify the relationship between FANCI and miR-218 expression.
Verification of FANCI expression by real time quantitative PCR (RT-qPCR)
We isolated total RNAs from LUAD tissues and reverse-transcribed them into complementary DNA (cDNA), and then used RT-qPCR to validate the expression of FANCI. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was amplified as an internal control. The primer sequences were as follows:
- FANCI-forward: CCACCTTTGGTCTATCAGCTTC.
- FANCI-reverse: CCACCTTTGGTCTATCAGCTTC.
- GAPDH-forward: GGAGCGAGATCCCTCCAAAAT.
- GAPDH-reverse: GGCTGTTGTCATACTTCTCATGG.
Human Protein Atlas (HPA) analysis
Immunohistochemistry (IHC) staining data were used from the HPA database to analyze the protein level of FANCI in LUAD samples. FANCI protein expression included four levels: high, medium, low, and undetected. The scoring system involved the percentage of positively-stained cells as well as the intensity of staining.
Exploring the relationship between the expression of FANCI and clinical features in TCGA database
We downloaded the clinical information and mRNA sequencing data of LUAD patients from the TCGA database, and the clinicopathological features and FANCI mRNA expression values of 475 patients with LUAD were obtained via screening. Patients were divided into a high and low group according to the median expression of FANCI. We analyzed the correlation between the expression of FANCI and tumor size, gender, lymph node metastasis, age, clinical stage, distant metastasis, and survival. The effects of clinicopathological features and FANCI expression on survival were analyzed by univariate and multivariate analyses.
Tumor Immune Estimation Resource (TIMER) analysis
The TIMER online tool (https://cistrome.shinyapps.io/timer/), which is a website that analyzes immune infiltration across multiple cancer types, was used to assess these infiltrates on the LUAD sample data. We explored the connection between FANCI and immune cells as well as their marker genes. The parameters were selected as follows: “survival”, cancer type: “LUAD”, gene type: “FANCI”, clinical: “All”, and immune infiltration: “Default”, and survival analysis was conducted to establish a multivariate Cox proportional risk model, which adopted a Cox proportional hazard model and used R package (https://www.rstudio.com/products/rpackages/) “survival” to fit.
Statistical analysis
We used GraphPad Prism 8.0 (Amercan, GraphPad Software, LLC) software to test the differential expression. Pearson correlation analysis was used to determine the relationship between the expression of FANCI and miR-218. The χ2 test was applied to analyze the relationship between FANCI expression and clinicopathological characteristics. We performed univariate and multivariate survival analysis using the Cox proportional hazards regression models. We used SPSS (Amercan, IBM) statistical software and RStudio (https://www.rstudio.com/) to analyze the clinical characteristics. A P value <0.05 was considered statistically significant.
Results
DEGs and microRNAs analysis
We obtained 2,708, 1,159, and 1,756 DEGs from the GSE40791, GSE27262, and GSE75037 datasets, respectively (Figure 1A-1C). Among them, 608 DEGs overlapped; 177 were upregulated, and 431 were downregulated (Figure 1). From the GSE63805 dataset, a total of 27 DEMs (16 upregulated and 11 downregulated) miRNAs were extracted (Figure 1D and Table S1). These miRNAs and genes were recognized as differentially expressed in LUAD samples compared with normal samples.
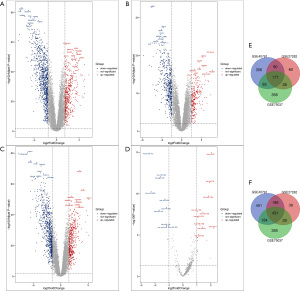
Functional enrichment analysis
The biological process that analyzed the overlapping DEGs included positive regulation of angiogenesis, positive regulation of cell-substrate adhesion, and extracellular matrix organization (Figure 2A). Regarding molecular functions (MF), it included heparin binding, calcium ion binding, and adenosine-triphosphate (ATP)-dependent microtubule motor activity (Figure 2B). Regarding cellular components (CC), it included extracellular space, proteinaceous extracellular matrix, and extracellular exosome (Figure 2C). Additionally, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways analyzed included malaria, extracellular matrix (ECM)-receptor interaction, and protein digestion and absorption (Figure 2D).

Regarding biological processes (BP) analysis of DEMs, the pathways were concentrated in the regulation of nucleobase, nucleoside, nucleotide, and nucleic acid metabolism (Figure 3A). In addition, the most enriched Gene Ontology (GO) terms in MF were transcription factor activity (Figure 3B). Regarding CC, the DEMs were concentrated in the actin cytoskeleton, cytoplasm, and nucleus (Figure 3C). As shown in Figure 3D, the biological pathway analysis included proteoglycan syndecan-mediated signaling events, the erythroblastic leukemia viral oncogene homolog (ERBB) receptor signaling network, the nectin adhesion pathway, plasma membrane estrogen receptor signaling, and signaling events mediated by the hepatocyte growth factor receptor.
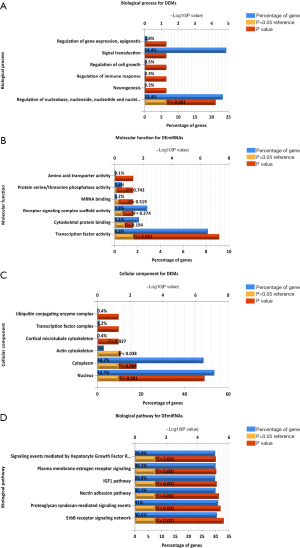
PPI network analysis
We used the STRING website to analyze the protein interactions of the overlapping genes. We constructed the network of DEGs, comprising 552 protein (Figure 4A), and used the MCODE Cytoscape plug-in to assess the functional modules. Module 1 was identified as consisting of 48 nodes and 1,075 edges, including FANCI, ANLN, GINS2, TPO2A, etc. (Figure 4B).
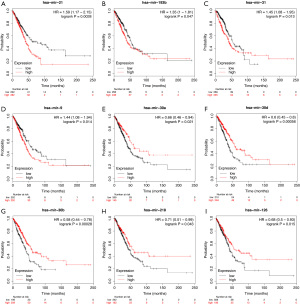
Kaplan-Meier analysis of DEMs
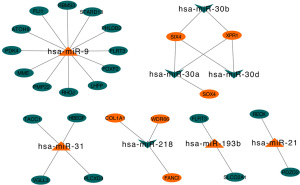
We used the KM plotter to predict the prognostic values of the 27 identified DEMs. The results showed that the high expression of miR-21, miR-193b, miR-31, and miR-9 (P<0.05) were associated with poor OS. Additionally, the low expression of miR-30a, miR-126, miR-30d, miR-218, and miR-30b (P<0.05) was related with worse OS (Figure 5).
DEM target mRNA prediction and miRNA-mRNA pairs
A total of 2,805 DEM target genes were predicted using the miRecord database, including 798 target genes predicted by downregulated DEMs and 2,007 target genes predicted by upregulated DEMs (Figure 6). These targets predicted by downregulated DEMs and upregulated DEMs intersected with upregulated DEGs and downregulated DEGs, respectively. Six upregulated DEGs were predicted by downregulated DEMs, and 20 downregulated DEGs were predicted by upregulated DEMs. A DEM-DEG regulatory network was constructed using Cytoscape (Figure 7).
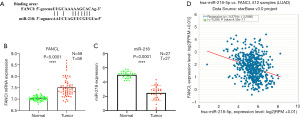
MiR-218 negatively regulates FANCI mRNA expression
TargetScan was used to predict that miR-218 could target FANCI, and the binding site of FANCI was identified in the sequence of miR-218 (Figure 8A). In addition, FANCI expression and miR-218 in LUAD samples was detected using GEO data, and the potential connection between FANCI and miR-218 was explored by the TargetScan database. Compared with normal lung samples, FANCI mRNA expression in LUAD tissues was significantly upregulated (Figure 8B), and miR-218 expression in LUAD samples was markedly downregulated (Figure 8C), which indicated that FANCI was negatively correlated with miR-218 (Figure 8D).
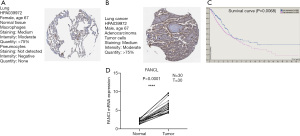
FANCI mRNA expression detection by qRT-PCR and protein expression by immunohistochemistry (IHC)
The IHC staining for FANCI in the HPA database was compared between LUAD tumor samples and normal lung samples using the HPA039972 antibody. FANCI was “undetected” in normal lung tissue (Figure 9A), and was detected as “moderate” in tumor samples (Figure 9B). By analyzing the correlation between FANCI expression and prognosis in the HPA database, a total of 696 high expression samples and 298 low expression samples were obtained. The OS was significantly lower in the FANCI high expression group (log-rank P=0.0068, Figure 9C). We also verified the FANCI expression using our own samples by RT-qPCR, and FANCI was notably high in LUAD samples (t=12.21, P<0.0001, Figure 9D).
Relationship between clinicopathological features and FANCI expression
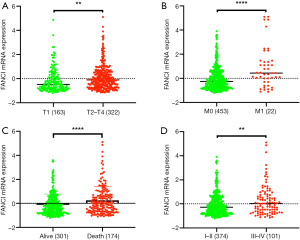
Based on TCGA database, we combined the clinicopathological data and found that FANCI was correlated with tumor size (χ2=13.96, P<0.001), distant metastasis (χ2=45.39, P<0.001), lymphatic metastasis (χ2=3.88, P<0.05), and stage (χ2=11.03, P<0.05, Table 1). Meanwhile, FANCI expression was significantly correlated with tumor size (P<0.01), distant metastasis (P<0.0001), survival status (P<0.0001), and tumor (T), regional lymph node (N), metastasis (M) and clinical stage (P<0.01, Figure 10). Furthermore, multivariate Cox analysis showed that FANCI was a significant independent factor for poor OS (P<0.05, Table 2). Based on TCGA database, the overall 5-year survival rate of the FANCI high expression group was poorer than the low expression group (P<0.01; Figure S1).
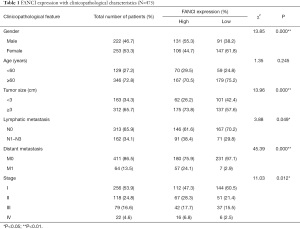
Full table
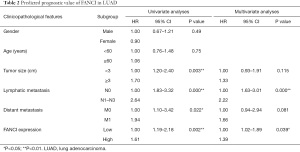
Full table
Relationship between FANCI and immune infiltrates
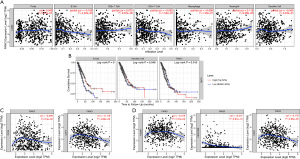
The TIMER website analysis showed that the expression of FANCI was associated with B cell and neutrophil infiltration in LUAD (P<0.05, Figure 11A). Combined with survival data, LUAD samples with high infiltration levels of B cells had a good prognosis compared to low B cell infiltration levels (P<0.05), and same results were observed in dendritic cells (P<0.05, Figure 11B). In order to further explore the relationship between FANCI and infiltrating immune cells, the relationship between FANCI expression and marker genes in tumor-infiltrating immune cells was evaluated. The results showed that FANCI expression was related to marker genes Cluster of Differentiation (CD) 19 and CD79A in B cells (Figure 11C). The marker genes of neutrophils were CECAM8, ITGAM, and CCR7 (Figure 11D).
Discussion
Cancer is a major global public health problem, and lung cancer has been the most common cause of cancer-related mortality in recent years (5). Standard treatment for lung cancer involves surgical resection, chemotherapy, radiotherapy, and immunotherapy. Despite these treatments, there has been no significant change in the OS rate of LUAD patients (6), and thus, finding new therapeutic targets for treatment is crucial. Through accurate microarray analysis of tumors, researchers can screen out biomarkers related to tumor occurrence and development for early diagnosis, therapeutic targets, and prognosis evaluation of cancer patients (7).
In the present study, the GO analysis results indicated that DEGs were primarily concentrated in the positive regulation of angiogenesis, positive regulation of cell-substrate adhesion, heparin binding, calcium ion binding, extracellular space, and proteinaceous extracellular matrix. These results are consistent with previously published findings that angiogenesis and cell substrate adherence are closely related to tumor invasion and metastasis (8,9). The KEGG analysis of the DEGs was mainly concentrated in ECM-receptor interaction, malaria, as well as protein digestion and absorption. In addition, previous studies have shown that ECM receptor interactions play a critical role in tumor progression (10). ECM is upregulated in prostate cancer tissues (11), and is involved in the metastasis of gastric cancer (12). ECM may also promote epithelial-mesenchymal transition (EMT) in colorectal cancer (13). In addition, ECM in the glioblastoma microenvironment is very important for the pathological characteristics of abnormal neovascularization and diffuse tumor cell infiltration (14). These previous studies indicate that ECM is involved in tumor progression and metastasis.
MicroRNAs (miRNAs) are small, noncoding RNA molecules (20–22 nucleotides in length) that regulate gene expression through translation inhibition or mRNA degradation (15). MiRNAs can act as both oncogenes and tumor suppressors by inhibiting the expression of target genes (16). Considerable research has shown that changes in the expression of miRNAs are associated with a variety of cancers, including lung cancer (17).
In total, 2,805 genes were targeted by DEMs that were associated with poor OS for LUAD patients, and 26 DEGs were obtained through the intersection of DEM-targeted genes with DEGs. These 26 DEGs and eight DEMs constituted 30 pairs of miRNA-mRNA regulatory relationships. Among the DEGs in these regulatory relationships, only FANCI was identified as the hub gene. Our bioinformatics prediction showed that miRNA-218 is closely related with FANCI, and may be an upstream miRNA that negatively regulates the mRNA expression of FANCI. Numerous studies have demonstrated that miR-218 is involved in the development of various types of cancer by targeting a single gene, and FANCI also plays an important role in many types of cancer. However, the fact that miR-218 negatively regulates FANCI mRNA expression in LUAD has not been reported previously.
MiR-218 has been shown to be downregulated in various malignancies, playing vital functions similar to tumor suppressor genes. Zhang et al. reported that miR-218 inhibits tumor angiogenesis in gastric cancer by targeting Roundabout Guidance Receptor 1 (ROBO1) (18). Furthermore, Ahmadinejad et al. demonstrated that the miR-218 is associated with poor prognosis in breast cancer (19). Xu et al. found that miR-218 inhibits LUAD progression by directly targeting BMI1 Proto-Oncogene, Polycomb Ring Finger (BMI-1) to regulate the EMT and phosphatidylinositol 3 kinase(PI3K)/protein kinase B (AKT) signaling pathways (20). Our finding that lower expression of miR-218 suppresses LUAD progression is consistent with these reports.
Fanconi anemia (FA) is caused by the mutation of the FA family protein, and can lead to chromosome instability and other diseases, including cancer (21,22). Abbasi et al. showed that FANCA was a breast cancer-susceptibility allele, which is related to familial breast cancer (23). Rockfield et al. reported that mutation of both FA Complementation Group D2 (FANCD2) was associated ovarian cancer (24). Zheng et al. emphasized that FANCI may act as an oncogene in LUAD and promote cell proliferation via the mitogen-activated protein kinase kinase (MEK)/extracellular regulated protein kinases (ERK)/matrix metalloproteinases (MMPs) pathway in collaboration with Inosine Monophosphate Dehydrogenase 2 (IMPDH2) (25).
In this study, bioinformatics analysis revealed that FANCI is the hub gene of LUAD, and was highly expressed in LUAD samples and associated with metastasis and poor prognosis. According to GEO and HPA databases, FANCI expression in LUAD tissues was higher than in the normal controls at both the mRNA and protein levels. Based on TCGA database, FANCI was correlated with tumor size, distant metastasis, lymphatic metastasis, and stage. In addition, FANCI has also been demonstrated to be an independent factor of LUAD patient prognosis, with higher expression indicating poorer prognosis.
Tumor-infiltrating immune cells in the lung cancer microenvironment can identify and also promote tumor cell apoptosis to inhibit tumor progression. Meanwhile, immune cells can screen tumor cells more suitable for survival in the microenvironment to promote tumor progression (26). A previous study has shown that FANCI is negatively correlated with dendritic cells, and the high expression of FANCI has fewer dendritic cells, leading to fewer T cells. T cell infiltration usually predicts better clinical outcomes in patients (27). In addition, the role of tumor-infiltrating B cells in the tumor microenvironment has also attracted increasing attention. Most studies have shown that B cell infiltration in lung cancer is associated with a good prognosis. This is consistent with the results of this study; the prognosis of high B cell infiltration in LUAD was better than that of low B cell infiltration (28). Our results also showed that the expression of FANCI was negatively correlated with B cells, indicating that the infiltration of B cells was less when FANCI was highly expressed, which may be one of the reasons for the poor prognosis of patients with high FANCI expression. In summary, changes in the immune microenvironment may be the mechanism through which FANCI leads to a poor prognosis in LUAD.
Our study obtained 608 DEGs and 26 DEMs, nine DEM-targeted DEGs, and 30 DEM-DEG pairs. The ECM-receptor interaction process may be related to LUAD progression. FANCI and its paired miRNA-218 may be critical genes for effective biomarkers to evaluate the poor prognosis of LUAD patients in a timely manner, and to provide potential therapeutic targets. Further analysis confirmed that FANCI was highly expressed in LUAD; we observed that higher expression of FANCI in LUAD is associated with larger tumor size, lymph node metastases, distant metastasis, higher grades, and poorer prognosis. We also identified that FANCI is an independent prognostic factor in LUAD. Therefore, miR-218 may negatively regulate FANCI, promoting metastasis, which leads to late stage and poor prognosis of LUAD by ECM-receptor interaction. The data of this study mainly comes from the published public database and belongs to the exploration of bioinformatics analysis. The deep relationship between miRNA/FANCI and lung adenocarcinoma still needs to be verified by later relevant in vivo and in vitro experimental studies.
This study showed that 608 DEGs and 26 DEMs were screened out, among which 30 pairs of DEMs composed of 26 DEGs and eight DEMs were identified. As the hub gene of LUAD, FANCI was highly expressed in LUAD. MiR-218 could target FANCI, and FANCI expression was negatively correlated with miR-218 expression, which may be the upstream miRNA negatively regulating the expression of FANCI mRNA. FANCI expression was found to be related to tumor size, lymphatic metastasis, distant metastasis, and stage, and was also an independent predictor of patient prognosis. Enrichment pathway analysis indicated that ECM receptor interaction may be closely related to the progression of LUAD. FANCI may be a key gene to determine the metastasis and poor prognosis in patients with LUAD, and is expected to be a new therapeutic target for LUAD. The existence of these targeted relationships requires further study, which may elucidate the mechanisms of LUAD metastasis and provide new methods for future diagnosis and treatment exploration.
Conclusions
As described above, we speculate that miR-218 may negatively regulate FANCI via ECM receptor interaction, leading to metastasis and poor prognosis of LUAD. FANCI may be a key gene to determine the metastasis and poor prognosis in patients with LUAD. Changes in the immune microenvironment may be the mechanism through which FANCI leads to a poor prognosis of LUAD.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-3823
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-3823
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-3823). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University (No. 2019-KY-255), and the patients and their families agreed to the tissue samples collection and their use in research.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Li H, Li QD, Wang MS, et al. Smoking and air pollution exposure and lung cancer mortality in Zhaoyuan County. Int J Hyg Environ Health 2013;216:63-70. [Crossref] [PubMed]
- Chen M, Liu X, Du J, et al. Differentiated regulation of immune-response related genes between LUAD and LUSC subtypes of lung cancers. Oncotarget 2017;8:133-44. [Crossref] [PubMed]
- Wang K, Chen R, Feng Z, et al. Identification of differentially expressed genes in non-small cell lung cancer. Aging (Albany NY) 2019;11:11170-85. [Crossref] [PubMed]
- Xu J, Li Y, Lu J, et al. The mRNA related ceRNA-ceRNA landscape and significance across 20 major cancer types. Nucleic Acids Res 2015;43:8169-82. [Crossref] [PubMed]
- Nasim F, Sabath BF, Eapen GA. Lung Cancer. Med Clin North Am 2019;103:463-73. [Crossref] [PubMed]
- Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet 2017;389:299-311. [Crossref] [PubMed]
- Tiwari M. Microarrays and cancer diagnosis. J Cancer Res Ther 2012;8:3-10. [Crossref] [PubMed]
- Tao X, Cheng L, Li Y, et al. Expression of CRYAB with the angiogenesis and poor prognosis for human gastric cancer. Medicine (Baltimore) 2019;98:e17799 [Crossref] [PubMed]
- Singh A, Winterbottom E, Daar IO. Eph/ephrin signaling in cell-cell and cell-substrate adhesion. Front Biosci (Landmark Ed) 2012;17:473-97. [Crossref] [PubMed]
- Cao L, Chen Y, Zhang M, et al. Identification of hub genes and potential molecular mechanisms in gastric cancer by integrated bioinformatics analysis. PeerJ 2018;6:e5180 [Crossref] [PubMed]
- Andersen MK, Rise K, Giskeødegård GF, et al. Integrative metabolic and transcriptomic profiling of prostate cancer tissue containing reactive stroma. Sci Rep 2018;8:14269. [Crossref] [PubMed]
- Yan P, He Y, Xie K, et al. In silico analyses for potential key genes associated with gastric cancer. PeerJ 2018;6:e6092 [Crossref] [PubMed]
- Rahbari NN, Kedrin D, Incio J, et al. Anti-VEGF therapy induces ECM remodeling and mechanical barriers to therapy in colorectal cancer liver metastases. Sci Transl Med 2016;8:360ra135 [Crossref] [PubMed]
- Cui X, Morales RT, Qian W, et al. Hacking macrophage-associated immunosuppression for regulating glioblastoma angiogenesis. Biomaterials 2018;161:164-78. [Crossref] [PubMed]
- Mishra S, Yadav T, Rani V. Exploring miRNA based approaches in cancer diagnostics and therapeutics. Crit Rev Oncol Hematol 2016;98:12-23. [Crossref] [PubMed]
- Zhao Q, Cao J, Wu YC, et al. Circulating miRNAs is a potential marker for gefitinib sensitivity and correlation with EGFR mutational status in human lung cancers. Am J Cancer Res 2015;5:1692-705. [PubMed]
- Yang X, Zhang Q, Zhang M, et al. Serum microRNA Signature Is Capable of Early Diagnosis for Non-Small Cell Lung Cancer. Int J Biol Sci 2019;15:1712-22. [Crossref] [PubMed]
- Zhang X, Dong J, He Y, et al. miR-218 inhibited tumor angiogenesis by targeting ROBO1 in gastric cancer. Gene 2017;615:42-9. [Crossref] [PubMed]
- Ahmadinejad F, Mowla SJ, Honardoost MA, et al. Lower expression of miR-218 in human breast cancer is associated with lymph node metastases, higher grades, and poorer prognosis. Tumour Biol 2017;39:1010428317698362 [Crossref] [PubMed]
- Xu L, Sun HB, Xu ZN, et al. MicroRNA-218 regulates the epithelial-to-mesenchymal transition and the PI3K/Akt signaling pathway to suppress lung adenocarcinoma progression by directly targeting BMI-1. Eur Rev Med Pharmacol Sci 2019;23:7978-88. [PubMed]
- Kelaidi C, Makis A, Petrikkos L, et al. Bone Marrow Failure in Fanconi Anemia: Clinical and Genetic Spectrum in a Cohort of 20 Pediatric Patients. J Pediatr Hematol Oncol 2019;41:612-7. [Crossref] [PubMed]
- Engel NW, Schliffke S, Schüller U, et al. Fatal Myelotoxicity Following Palliative Chemotherapy With Cisplatin and Gemcitabine in a Patient With Stage IV Cholangiocarcinoma Linked to Post Mortem Diagnosis of Fanconi Anemia. Front Oncol 2019;9:420. [Crossref] [PubMed]
- Abbasi S, Rasouli M. A rare FANCA gene variation as a breast cancer susceptibility allele in an Iranian population. Mol Med Rep 2017;15:3983-8. [Crossref] [PubMed]
- Rockfield S, Kee Y, Nanjundan M. Chronic iron exposure and c-Myc/H-ras-mediated transformation in fallopian tube cells alter the expression of EVI1, amplified at 3q26.2 in ovarian cancer. Oncogenesis 2019;8:46. [Crossref] [PubMed]
- Zheng P, Li L. FANCI Cooperates with IMPDH2 to Promote Lung Adenocarcinoma Tumor Growth via a MEK/ERK/MMPs Pathway. Onco Targets Ther 2020;13:451-63. [Crossref] [PubMed]
- Merlo A, Dalla Santa S, Dolcetti R, et al. Reverse immunoediting: When immunity is edited by antigen. Immunol Lett 2016;175:16-20.
- Zhang J, Endres S, Kobold S. Enhancing tumor T cell infiltration to enable cancer immunotherapy. Immunotherapy 2019;11:201-13. [Crossref] [PubMed]
- Wang SS, Liu W, Ly D, et al. Tumor-infiltrating B cells: their role and application in anti-tumor immunity in lung cancer. Cell Mol Immunol 2019;16:6-18. [Crossref] [PubMed]
(English Language Editor: A. Kassem)


