
TP53 and CDKN2A mutations in patients with early-stage lung squamous cell carcinoma: an analysis of the correlations and prognostic outcomes
Introduction
Lung squamous cell carcinoma (LUSC) accounts for 20–30% of non-small-cell lung cancers (NSCLCs) and results in approximately 400,000 deaths annually in the United States. Unfortunately, to date, very few personalized therapies have been developed for LUSC due to the limited understanding of the molecular targets (1). Previous profiling efforts have demonstrated that mutations in tumor protein p53 (TP53) represent the most frequent (81%) genomic alteration found in LUSC (2). TP53 encodes the tumor suppressor protein p53, binds directly to chromatin in the nucleus, and plays an important role in the regulation of the cell cycle, apoptosis, autophagy, and DNA repair in response to oncogenic stress (3). The position, nature, and functional effects of mutations on protein structure and activity have led to a recent classification of TP53 mutations, and it is now recognized that various classes of mutations have differential prognostic effects. However, data on the prognostic or predictive effects of TP53 status in NSCLC are limited and inconclusive (4). To date, there is still a paucity of drugs approved for targeting TP53 mutations in cancer patients, and the prognostic value of TP53 in early-stage LUSC is unclear. Thus, this study examined the prognostic value of TP53 in early-stage LUSC.
Cyclin dependent kinase inhibitor 2A (CDKN2A), a known tumor suppressor gene that encodes the p16INK4A and p14ARF proteins, is inactivated in 72% of LUSC cases (2). Patients who are carriers of certain CDKN2A mutations show increased risks of malignant neoplasms, particularly pancreatic, lung, and head and neck cancers (5). LUSC is characterized by frequent TP53 mutations and CDKN2A alterations (2,6). Previous studies have reported that the degradation of the p53 protein by the ubiquitin pathway is mediated by its binding to mouse double minute 2 (MDM2). However, the expression of MDM2 mRNA and protein is negatively regulated by p14ARF in the nucleus (7,8). While these alterations have increased our understanding of the molecular pathology of LUSC, the impact of TP53/CDKN2A status on the clinical outcomes of patients with early-stage LUSC is unclear.
This study analyzed the mutational landscape of 16 early-stage, surgically resected LUSC patients using targeted next-generation sequencing (NGS) encompassing 59–1,021 cancer-related genes. Furthermore, we utilized a well annotated specimen set that permits analysis of mutations, alone or in combination, with outcome. This study aimed to evaluate the association of TP53 and CDKN2A status, as well as the prognostic value of these two genes combined in early-stage, surgically resected LUSC. We present the following article in accordance with the REMARK reporting checklist (available at https://dx.doi.org/10.21037/atm-21-3709).
Methods
Patients and samples
Sixteen early-stage, surgically resected LUSC samples were obtained from the Fujian Cancer Hospital in Fuzhou, China, from August 2018 to August 2019. All patients provided written informed consent and received NGS testing at the Geneplus-Beijing Institute. NGS testing covered approximately 1.4 Mbp genomic regions of 1,021 cancer-related genes (or approximately 230 Kbp genomic regions of 59 genes for some patients) (Table S1). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by regional ethics board of Fujian Cancer Hospital (No.: SQ2020-055-01) and informed consent was taken from all the patients.
Gene expression databases
Information regarding TP53 and CDKN2A alterations and survival times in patients with LUSC was downloaded from The Cancer Genome Atlas (TCGA), an open access database that is publicly available at http://www.cbioportal.org. The Lung Squamous Cell Carcinoma (TCGA, Firehose Legacy), Lung Squamous Cell Carcinoma (TCGA, Nature 2012), and Lung Squamous Cell Carcinoma (TCGA, PanCancer Atlas) datasets were selected as the data source as they contained only early-stage, surgically resected LUSC samples. There was a total of 1176 LUSC samples (available online: https://cdn.amegroups.cn/static/public/atm-21-3709-1.xlsx). The gene set of interest, “TP53 CDKN2A”, was entered in the input box. Mutation and survival data were downloaded from the cBioPortal website after submitting the query regarding “TP53 CDKN2A” in the input box. Data were merged according to the unique patient ID, such as “TCGA-18-3406-01”. Altogether, 841 pieces of mutation data and 979 pieces of survival data were downloaded. Analysis of the data revealed 349 pieces of duplicated patient data, which were discarded. The duplication was largely due to overlapping data with another selected study. After the merge, there were 492 pieces of data from early-stage LUSC patients. Each piece of data contained the mutation type of CDKN2A and TP53 as well as the survival time of the patient. The TP53 and CDKN2A mutations were divided into different groups based on the different exons containing the mutations. However, due to limited information from the cBioPortal database, there were 13 cases without mutation counts in TP53-wild-type patients and 10 cases without mutation counts in TP53-mutated patients. No statements of approval or informed consent were required for this section of the study, as all data was obtained from an open access database.
Statistical methods
Fisher’s exact test and the Mann-Whitney test were utilized to analyze the categorical and continuous variables. Survival curves were analyzed using the Kaplan-Meier method and log-rank tests. The Cox proportional hazards model was used to evaluate associations between clinicopathological characteristics and patient survival. Overall survival (OS) and disease-free survival (DFS) data were obtained from the cBioPortal website directly. Statistical analyses were performed using GraphPad Prism 5.0. The statistical significance (alpha-value) threshold was fixed at 0.05, and all P values were three-sided.
Results
TP53 and CDKN2A exhibited a higher frequency of somatic mutations than other cancer-related genes
A retrospective study was conducted on 16 LUSC patients involving genomic profiling via targeted NGS encompassing 59–1,021 cancer-related genes. Among LUSC patients, the rate of TP53 mutation was 87.5% (14/16), while the rate of CDKN2A mutation was 43.8% (7/16). Interestingly, CDKN2A mutations were accompanied by TP53 mutations. TP53 and CDKN2A were among the most frequently mutated genes, whereas F-box and WD repeat domain containing 7 (FBXW7), notch receptor 4 (NOTCH4), epidermal growth factor receptor (EGFR), BRCA1 DNA repair associated (BRCA1) had lower mutation rates in our study cohort (Figure 1A). The mutation frequencies of TP53, NOTCH4, kelch like ECH associated protein 1 (KEAP1), phosphatase and tensin homolog (PTEN), ROS proto-oncogene 1, receptor tyrosine kinase (ROS1), EGFR, and BRCA1 were comparable with those in the TCGA data (Figure 1B). Patients with early-stage LUSC exhibited higher TP53 and CDKN2A mutation frequencies compared to other cancer-related genes.
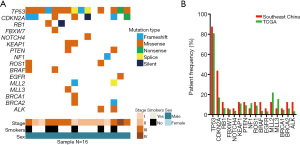
TP53 mutation and CDKN2A mutation profiling and patient characteristics
In the 492 early-stage LUSC patients, the mutation rate of TP53 was 83.13% (409/492). Exons 4–8 were the most frequent mutation sites for TP53, accounting for 66.9% of all mutations (329/492). Exons 9 and 10 were rarely mutated, accounting for 2.4% of all mutations (12/492). Multiple mutations occurred in 4.5% of patients (22/492), and 9.3% (46/492) of mutations could not be classified (Figure 2A,2B). TP53 mutations, mainly missense mutations, were the most common mutations in early-stage LUSC (Figure 2C).
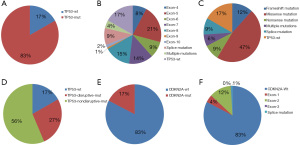
In another classification, TP53 mutation status was divided into wild type, disruptive mutations, and nondisruptive mutations, as previously described (9). A total of 131 patients (27%) had TP53 disruptive mutations, and 278 patients (56%) showed nondisruptive TP53 mutations (Figure 2D). Patient and tumor characteristics based on TP53 status are shown in Table S2. Dual TP53/CDKN2A mutations were observed in 78 patients (15.8%). CDKN2A mutation status was associated with TP53 mutation status (P=0.040).
Of the 492 patients, 406 patients did not have CDKN2A mutations, 21 patients had exon 1 mutations, 60 had exon 2 mutations, 1 had exon 3 mutations, and 4 had splice mutations (Figure 2E,2F). Patient and tumor characteristics based on CDKN2A status are shown in Table S3. There were no statistically significant differences in tumor characteristics.
Kaplan-Meier analyses of survival time according to TP53 status in early-stage LUSC
Consistent with the prognostic capacity of tumor staging, among the entire cohort of 492 patients, the Kaplan-Meier survival curve indicated that patients with different tumor stages had significantly different OS times (P=0.009; Figure 3A). Similarly, there was a significant difference in OS based on TP53 mutant or wild-type status with distinct tumor staging (P=0.020 and P=0.025, respectively) (Figure 3B,3C). However, the year of initial diagnosis did not significantly affect prognosis (P=0.595; Figure S1A).
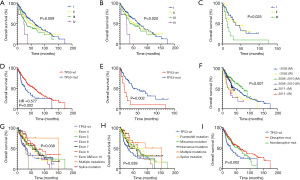
Furthermore, TP53 mutation was a positive prognostic factor for OS and DFS in early-stage LUSC (Figure 3D and Figure S1B). The estimated OS times for patients with wild-type TP53 and mutated TP53 were 28.94 months and 60.48 months, respectively [hazard ratio (HR) 0.577; 95% confidence interval (CI), 0.390 to 0.878; P=0.002]. This prompted us to investigate the association between patient survival time and tumor stage as well as TP53 mutations. In stage III patients, OS was affected by TP53 status (HR 4.21; 95% CI, 1.68 to 10.56; P=0.002 for OS), but no significant difference was identified in stage I–II patients (Figure 3E and Figure S1C,S1D). In addition, OS for the entire cohort was influenced by the year of initial diagnosis according to TP53 status (P =0.007; Figure 3F).
TP53 mutations were divided according to the affected exons. Patients with different mutated exons had significantly different OS times (P=0.038; Figure 3G). However, the difference in DFS was not statistically significant (Figure S1E). Diverse types of TP53 mutations can occur, and the TP53 mutation type can affect the prognosis of LUSC patients (P=0.045 for OS and P=0.039 for PFS; Figure 3H and Figure S1F). In another classification, TP53 mutation status was divided into wild type, disruptive mutations, and nondisruptive mutations. The difference in survival between these mutation groups was also statistically significant (P=0.002 for OS and P=0.039 for PFS; Figure 3I).
Survival analysis of TP53 and CDKN2A status in early-stage LUSC
Few studies have demonstrated the prognostic impact of CDKN2A mutations in early-stage LUSC. The results demonstrated that the CDKN2A mutation was a negative prognostic factor in early-stage LUSC (Figure 4A). The estimated OS times for patients with wild-type CDKN2A and mutated CDKN2A were 62.81 months and 37.55 months, respectively (HR 0.692; 95% CI, 0.479 to 0.998; P=0.026). DFS was not influenced by CDKN2A status (HR 0.823; 95% CI, 0.596 to 1.138; P=0.209; Figure S2A). The mutation type was further divided according to the affected exons, and the difference between exon groups was statistically significant for OS (P=0.024; Figure 4B). Again, DFS was not significantly different between the exon groups (P=0.197; Figure S2B). The patients were further divided into different groups based on the mutation type of CDKN2A (wild type or mutated) and TP53 (wild type or mutated). The results revealed that the survival of the 4 different groups was significantly different (P<0.001 for OS and DFS; Figure 4C and Figure S2C). Patients with TP53 mutated/CDKN2A wild-type status showed longer OS and DFS compared to patients in the other 3 groups. This suggested that TP53 mutation and CDKN2A mutation types are prognostic factors in early-stage LUSC.
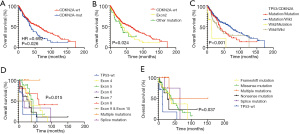
The prognostic value of CDKN2A and TP53 mutation types was further investigated. Survival curves of CDKN2A-mutated patients indicated that TP53 wild-type patients had a poor prognosis (P=0.015 for different mutation sites; P=0.037 for different mutation types; Figure 4D,4E). However, in CDKN2A wild-type patients, OS was not influenced by TP53 status (P=0.219 for different mutation sites in different exons; P=0.154 for different mutation types; Figure S2D,S2E). Interestingly, when TP53 mutation status was divided into wild type, disruptive mutations, and nondisruptive mutations, OS was not influenced by CDKN2A status (Figure S2F,S2G).
Correlation of mutation counts and survival time
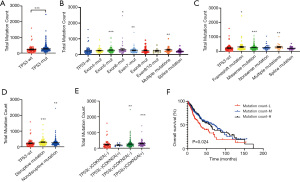
Oncogenic stress triggers the DNA damage response which involves p53-mediated DNA repair to trigger cell cycle arrest and cell death by apoptosis or senescence (10). When TP53 is mutated, more mutations may occur. The association between mutation counts during early-stage LUSC and at different tumor stages was investigated. Different stages exhibited similar mutation frequencies. Total mutation counts were not influenced by stage regardless of TP53 status (Figure S3A-S3C). Interestingly, patients with mutated TP53 harbored more total mutations (Figure 5A). However, the CDKN2A wild-type group and CDKN2A-mutated group exhibited similar mutation counts (Figure S3D). In addition to the exon 4 mutation, different TP53 mutation sites were related to higher mutation counts compared to wild-type TP53 (Figure 5B). Similarly, patients with TP53 frameshift mutation, missense mutation, and multiple mutations had more mutations, with the exception of splice mutations and nonsense mutations (Figure 5C). In addition, patients with disruptive and nondisruptive TP53 mutations all presented with higher mutation counts than TP53 wild-type patients (Figure 5D). Moreover, compared to the TP53 and CDKN2A wild-type cohorts, the TP53 mutation cohort had more mutations irrespective of CDKN2A mutation status. These results suggested that mutation count is associated with TP53 status, independent of CDKN2A status (Figure 5E). The number of mutations was divided into 3 cohorts, namely, the low mutation count cohort, which included patients with 1–150 mutations; the medium mutation count cohort, which included patients with 151–300 mutations; and the high mutation count cohort, which included patients with over 301 mutations. OS was significantly shorter in the low mutation count cohort compared to patients in the medium and high mutation count groups (P=0.024; Figure 5F).
Discussion
In LUSC, recurrent mutations of TP53, FGFR1, FGFR2, FGFR3, DDR2, and genes of the PI3K pathway have been detected, as have quantitative gene abnormalities of PTEN and CDKN2A (1,6). This current study reviewed 16 patients with surgically resected LUSC and identified that TP53 and CDKN2A exhibited a higher frequency of somatic mutations than other cancer-related genes. These results were compared with those from the TCGA dataset, which is mainly composed of the Western population. Therefore, it is essential to further elucidate the association of TP53 status and CDKN2A status, as well as the prognostic value of these two genes together in early-stage, surgically resected LUSC patients in the Chinese population .
TP53 has been shown to be one of the most frequently mutated genes in lung cancers irrespective of histological type, with the vast majority of mutations clustering in exons 4 to 8 (11), which is consistent with our study. In addition, similar to the results of previous studies (3), missense mutations were the most common mutations observed in our early-stage LUSC cohort. In contrast to other studies (11), we demonstrated that TP53 mutations were not significantly associated with age or stage. As we only analyzed the influence of TP53 mutations in relation to early-stage, surgically resected LUSC rather than NSCLC, this may account for the difference observed between studies. Interestingly, CDKN2A mutation status was shown to be associated with TP53 mutation status, and in fact, CDKN2A mutations were present in 17% of early-stage LUSCs. However, there was no significant relationship between CDKN2A mutation status and tumor characteristics.
To date, data on the prognostic or predictive effect of TP53 in NSCLC have been limited and inconclusive. In a study cohort of 35 patients with NSCLC from a prospective phase II trial, TP53 mutation was predictive of resistance to induction therapy (cisplatin/etoposide plus radiation) (12). However, Schiller et al. failed to identify prognostic or predictive value in 197 patients with completely resected tumors enrolled in a randomized trial of postoperative radiotherapy plus chemotherapy (13). Negative results were also observed in JBRonchus (JBR), a randomized trial of patients with stage IB and II NSCLC assigned to treatment with cisplatin-based adjuvant chemotherapy (ACT) versus observation (OBS) (14). Another randomized trial of ACT versus OBS in patients with stage I to III NSCLC, the International Adjuvant Lung Cancer Trial (IALT), showed that TP53 mutation was neither prognostic nor predictive for OS after 8 years of follow-up. Ma et al. performed a pooled analysis of four randomized trials of ACT versus OBS and reported that TP53 mutation had no prognostic effect but was marginally predictive for survival from ACT (4).
This current investigation examined the prognostic value of TP53 in early-stage LUSC. Analysis of the TCGA data revealed a trend towards decreased OS with progressing tumor stage, regardless of TP53 status. Our study indicated that TP53mutation is a favorable prognostic factor in early-stage LUSC patients. This effect was only significant in stage III patients and not in stage I–II patients. OS was also significantly affected by the year of initial diagnosis, especially before 2010, and TP53 status. This discrepancy might be attributable to the development of ACT for use after surgery. TP53 mutations should be considered not only in terms of mutation status but also in terms of mutation site and mutation type. We found that the TP53 mutation site and mutation type were clinically meaningful. Similar to previous studies (15), patients with TP53 exon 4 or exon 6 mutations demonstrated poorer prognosis compared to patients with TP53 exon 5, exon 7, or exon 8 mutations. In addition, patients with multiple mutations demonstrated better prognosis than those with nonsense mutations. The former study divided TP53 mutation types into disruptive and nondisruptive, and found that nondisruptive mutations of TP53 are an independent prognostic factor of shorter survival time in EGFR-mutated NSCLC (16). However, our study showed that disruptive mutation of TP53 seemed to confer a longer survival time in early-stage LUSC, in agreement with Hou et al. (17).
CDKN2A alterations are frequent in all lung cancer expression subtypes (6). However, few reports have investigated the predictive or prognostic significance of CDKN2A in NSCLC. Our study indicated that CDKN2A mutations in early-stage LUSC are significantly associated with poor survival time. This was also the first study to analyze the association between TP53 status and CDKN2A status in early-stage, surgically resected LUSC patients. Patients with mutated TP53 and wild-type CDKN2A demonstrated a longer survival time compared with other early-stage LUSC patients. When CDKN2A status was divided into wild-type and mutated groups, survival curves of the mutated CDKN2A group showed that TP53 wild-type patients had a poorer prognosis. There were no significant differences between wild-type CDKN2A and TP53 status in terms of OS. The results suggested that CDKN2A mutation is a vital indicator for prognostic assessment according to TP53 status.
This investigation demonstrated that patients with TP53 mutations have longer OS and DFS among early-stage LUSC patients. Patients with TP53 mutation had more total mutations than those with wild-type TP53. Specifically, patients with different TP53 mutation sites and mutation types harbored different mutation counts and had higher mutation counts than those with wild-type TP53. Interestingly, patients with higher mutation counts had a longer survival time, which was consistent with the results demonstrating that patients with TP53 mutations had a longer survival time. Previous reports have suggested that the measurement of mutation counts is representative of tumor mutation burden (TMB) (18,19). Tumors with high TMB are thought to express more cancer-specific antigens (neoantigens) that can be recognized by the immune system (20). In the present study, data from the TCGA database included information on patients with early-stage, surgically resected LUSC from 1992 to 2013 who had good performance status and an active immune system. Moreover, patients with TP53 mutations diagnosed before 2010 who accepted limited therapy after surgery had a longer survival time than P53-wild-type patients. This discrepancy might be caused by differences in stage III patients and stage I–II patients. As resectable stage III LUSC has more circulating tumor cells, and patients with TP53 mutations may harbor higher mutation counts and express more neoantigens which can be recognized by the immune system compared to patients without TP53 mutations.
There were several limitations to this investigation including the small sample size of the cohort and inadequate information from the cBioPortal database. Future work should verified these results using other cohorts, such as data from the TCGA cohort. Further investigations regarding TP53 and CDKN2A mutations, and the prognosis of LUSC patients are required to fully evaluate the role of TP53/CDKN2A status as a prognostic and predictive variable in patients with LUSC. Though many biochemical aspects of p53 and CDKN2A regulation and activity were elucidated and it have demonstrated their inhibition of tumorigenesis (21,22), p53 and CDKN2A mutants differ considerably in form and function need furthermore investigation. We hope that different treatment strategies were adopted according to TP53 and CDKN2A status.
Acknowledgments
Funding: This work was supported by funding from a specific research fund for the public service sector, the National Natural Science Foundation of China (Grant Number 82002497), and the Science and Technology Program of Fujian Province (Grant Number 2020J05072).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-3709
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-3709
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-3709). The authors report funding support from the National Natural Science Foundation of China (Grant Number 82002497), and the Science and Technology Program of Fujian Province (Grant Number 2020J05072) for the article processing charges. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by regional ethics board of Fujian Cancer Hospital (No.: SQ2020-055-01) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Choi M, Kadara H, Zhang J, et al. Mutation profiles in early-stage lung squamous cell carcinoma with clinical follow-up and correlation with markers of immune function. Ann Oncol 2017;28:83-9. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. [Crossref] [PubMed]
- Bykov VJN, Eriksson SE, Bianchi J, et al. Targeting mutant p53 for efficient cancer therapy. Nat Rev Cancer 2018;18:89-102. [Crossref] [PubMed]
- Ma X, Le Teuff G, Lacas B, et al. Prognostic and Predictive Effect of TP53 Mutations in Patients with Non-Small Cell Lung Cancer from Adjuvant Cisplatin-Based Therapy Randomized Trials: A LACE-Bio Pooled Analysis. J Thorac Oncol 2016;11:850-61. [Crossref] [PubMed]
- Helgadottir H, Höiom V, Tuominen R, et al. Germline CDKN2A Mutation Status and Survival in Familial Melanoma Cases. J Natl Cancer Inst 2016; [Crossref] [PubMed]
- Testa U, Castelli G, Pelosi E. Lung Cancers: Molecular Characterization, Clonal Heterogeneity and Evolution, and Cancer Stem Cells. Cancers (Basel) 2018;10:248. [Crossref] [PubMed]
- Wang J, Ding S, Duan Z, et al. Role of p14ARF-HDM2-p53 axis in SOX6-mediated tumor suppression. Oncogene 2016;35:1692-702. [Crossref] [PubMed]
- Heo SH, Kwak J, Jang KL. All-trans retinoic acid induces p53-depenent apoptosis in human hepatocytes by activating p14 expression via promoter hypomethylation. Cancer Lett 2015;362:139-48. [Crossref] [PubMed]
- Poeta ML, Manola J, Goldwasser MA, et al. TP53 mutations and survival in squamous-cell carcinoma of the head and neck. N Engl J Med 2007;357:2552-61. [Crossref] [PubMed]
- Soussi T, Wiman KG. TP53: an oncogene in disguise. Cell Death Differ 2015;22:1239-49. [Crossref] [PubMed]
- Ma X, Rousseau V, Sun H, et al. Significance of TP53 mutations as predictive markers of adjuvant cisplatin-based chemotherapy in completely resected non-small-cell lung cancer. Mol Oncol 2014;8:555-64. [Crossref] [PubMed]
- Kandioler D, Stamatis G, Eberhardt W, et al. Growing clinical evidence for the interaction of the p53 genotype and response to induction chemotherapy in advanced non-small cell lung cancer. J Thorac Cardiovasc Surg 2008;135:1036-41. [Crossref] [PubMed]
- Schiller JH, Adak S, Feins RH, et al. Lack of prognostic significance of p53 and K-ras mutations in primary resected non-small-cell lung cancer on E4592: a Laboratory Ancillary Study on an Eastern Cooperative Oncology Group Prospective Randomized Trial of Postoperative Adjuvant Therapy. J Clin Oncol 2001;19:448-57. [Crossref] [PubMed]
- Tsao MS, Aviel-Ronen S, Ding K, et al. Prognostic and predictive importance of p53 and RAS for adjuvant chemotherapy in non small-cell lung cancer. J Clin Oncol 2007;25:5240-7. [Crossref] [PubMed]
- Jiao XD, Qin BD, You P, et al. The prognostic value of TP53 and its correlation with EGFR mutation in advanced non-small cell lung cancer, an analysis based on cBioPortal data base. Lung Cancer 2018;123:70-5. [Crossref] [PubMed]
- Canale M, Petracci E, Delmonte A, et al. Impact of TP53 Mutations on Outcome in EGFR-Mutated Patients Treated with First-Line Tyrosine Kinase Inhibitors. Clin Cancer Res 2017;23:2195-202. [Crossref] [PubMed]
- Hou H, Qin K, Liang Y, et al. Concurrent TP53 mutations predict poor outcomes of EGFR-TKI treatments in Chinese patients with advanced NSCLC. Cancer Manag Res 2019;11:5665-75. [Crossref] [PubMed]
- Oh JH, Jang SJ, Kim J, et al. Spontaneous mutations in the single TTN gene represent high tumor mutation burden. NPJ Genom Med 2020;5:33. [Crossref] [PubMed]
- Endris V, Buchhalter I, Allgäuer M, et al. Measurement of tumor mutational burden (TMB) in routine molecular diagnostics: in silico and real-life analysis of three larger gene panels. Int J Cancer 2019;144:2303-12. [Crossref] [PubMed]
- Zhang H, Deng YM, Chen ZC, et al. Clinical significance of tumor mutation burden and DNA damage repair in advanced stage non-small cell lung cancer patients. Eur Rev Med Pharmacol Sci 2020;24:7664-72. [PubMed]
- Hernández Borrero LJ, El-Deiry WS. Tumor suppressor p53: Biology, signaling pathways, and therapeutic targeting. Biochim Biophys Acta Rev Cancer 2021;1876:188556 [Crossref] [PubMed]
- Adib E, Nassar AH, Akl EW, et al. CDKN2A Alterations and Response to Immunotherapy in Solid Tumors. Clin Cancer Res 2021;27:4025-35. [Crossref] [PubMed]
(English Language Editor: J. Teoh)


