
Circular RNAs as potential regulators in bone remodeling: a narrative review
Introduction
Mammalian bone development starts with forming the fetus bone, which is accomplished by endochondral and intramembranous ossification (1). Postnatally, modeling and remodeling are responsible for the continuous changes in the size and shape of bones (2). Bone modeling deposits new bone without proceeding resorption; however, bone remodeling is a dynamic process that includes two opposite but mutually communicated parts of bone physiology: anabolic and catabolic activities (3). Traditionally, the process of bone remodeling has been divided into the following 4 sequential phases: (I) the activation phase in which hematopoietic progenitors are recruited to the damaged bone site; (II) the resorption phase in which mature osteoclasts secrete acid and enzymes to resorb bone matrix; (III) the reversal phase in which the apoptosis of osteoclasts is triggered, and osteoblast progenitors are recruited; and (IV) the formation phase in which osteoid is formed and subsequently mineralized (4,5). The coupling of these phases is controlled by osteoblast lineage cells, osteoclast lineage cells, and various regulatory factors in a precise spatiotemporal sequential manner (6). Skeletal disorders may occur if homeostasis is disturbed (7).
Circular ribonucleic acids (circRNAs), the newly discovered non-coding RNAs, have been the subject of extensive research in different types of diseases, such as lung cancers, neurological disorders, and cardiovascular diseases (8-10). Numerous studies have shown that circRNAs are pivotal regulators in bone remodeling. Zhang et al. reported that circRNAs were differentially expressed in human bone marrow stem cells (hBMSCs) after 7 days of osteogenic induction (11). Another study conducted a microarray analysis to detect the expression profiles of circRNAs during the osteoclastogenesis of RAW264.7 cells and found that several circRNAs were upregulated or downregulated in the three different stages of osteoclasts (i.e., pre-osteoclasts, mature osteoclasts, and activated osteoclasts) (12).
In the present review, we focus on the current detected molecular targets of circRNAs in bone remodeling and related disorders that are mainly driven by osteoblasts and osteoclasts. Three English and three Chinese databases [i.e., PubMed, Embase, MEDLINE (via Ovid), Chinese Biomedical Literature, China National Knowledge Infrastructure, and VIP databases] were searched to June 2021 without language restrictions. Studies exploring the roles of circRNAs in key bone remodeling mediators, such as Smad-dependent bone morphogenetic protein (BMP)/transforming growth factor beta (TGF-β), Wnts, runt-related transcription factor (RUNX), forkhead boxes (FOXs), colony-stimulating factor 1 (CSF-1), receptor activator of nuclear factor kappa B ligand (RANKL)/osteoprotegerin (OPG), and circRNA-related bone-remodeling disorders, were included. We present the following article in accordance with the Narrative Review reporting checklist (available at https://dx.doi.org/10.21037/atm-21-2114).
The biogenesis, characteristics, and functions of circRNAs
CircRNAs, unlike their linear cognate RNAs, are commonly generated by precursor messenger RNA (pre-mRNA) back splicing (13,14). The biogenesis of circRNA can be described as a covalent link between a downstream 3' donor site and an upstream 5' acceptor site by a 3'-5' phosphodiester bond (tail to head) that finally circularizes to a closed-loop. The mechanisms of circRNA biogenesis remain elusive; however, the following elements have been introduced to interpret the process (15,16) (see Figure 1A). (I) the reverse complementary sequence (RCS) flanking circRNA-derived exon: The base pairing of RCSs makes it easier for the donor site and the acceptor site to contact each other, thus favoring the back-splicing process; (II) RNA binding proteins (RBPs): RBPs, serving as trans-acting factors, form intronic paired RNAs to facilitate circRNA biogenesis in a similar way as RCSs; (III) the lariat from “exon skipping”: under this alternative form of splicing, called “exon skipping”, pre-mRNA fractions can be spliced to form lariat, some of which develop into mature circRNAs. Based on the inclusion or exclusion of different gene fractions, mature circRNAs are divided into the following three types: (I) exonic circular RNAs (ecRNAs); (II) circular intronic RNAs (ciRNAs); and (III) exon-intron circular RNAs (EIciRNAs) (17,18). Current detection results indicate that ecRNAs account for the highest proportion of circRNAs (19).
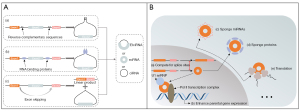
CircRNAs were discovered decades ago and are generally viewed as useless by-products (20,21). In 2013, Hansen et al. described the sponge effect of circRNAs toward microRNAs (miRNAs) in detail (22), and Memczak et al. described the widespread distribution of circRNAs in eukaryotic transcriptomes based on the sequencing and analysis of human, mouse, and nematode RNAs (23). Since then, the functions of circRNAs in diverse physiological and pathological processes have been investigated extensively. We summarize the recently discovered functions of circRNAs in Figure 1B, including the miRNA sponge role observed in the majority of circRNAs. A subset of circRNAs, which present with internal ribosome entry site (IRES) elements, has been associated with translating ribosomes and generating proteins in both experimental and endogenous contexts (24-26). Additionally, circRNAs have been shown to be scaffolds to sequester proteins (14); however, research in this area is limited.
Interestingly, circRNAs localized in the nucleus have very different roles to the above-mentioned cytoplasmic circRNAs. The biogenesis of some circRNAs was found to modulate their linear splicing process by competing for splice sites with their linear counterparts (27). Li et al. reported that EIciRNAs act as cis elements to regulate the expression levels of their parental genes by interacting with U1 small nuclear ribonucleoproteins and the Pol II transcription complex (28). Research has shown that circRNAs function as pivotal regulators in various physiological and pathological activities. Further, their circular conformation endows them with a unique ribonuclease-resistant character not possessed by their linear counterparts, making them ideal biomarkers and treatment targets (10).
CircRNAs modulate osteoblasts and bone formation
Osteoblasts are derived from bone marrow stem cells (BMSCs), and evidence has shown that circRNAs regulate the osteogenic differentiation process epigenetically (11). Additionally, tissue engineering gives rise to the possibility that other sources of stem cells could differentiate into osteoblasts. Adipose-derived stem cells (ASCs), which are more abundant and easily available than hBMSCs, are usually seen as suitable candidates for osteogenic induction. Microarray analysis was undertaken of human and mouse ASCs that had been divided into induced and non-induced groups, respectively, and the results showed that hundreds of circRNAs were differentially expressed between those two groups (29,30). Due to the excellent odontogenic and osteogenic potential, dental stem cells, such as human stem cells from apical papilla (hSCAP), human periodontal ligament stem cells (hPDLSCs), and human dental pulp stem cells (hDPSCs) represent promising seed cells for regenerative medicine. Their differentiation processes were recently shown to be correlated to the expression patterns of circRNAs (31-33).
To clarify their underlying mechanisms, we discuss some examples of circRNAs involved in regulating crucial osteogenic signaling pathway factors, including Smad-dependent BMP/TGF-β, Wnts, RUNX and FOXs (see Table 1).
Table 1
| CircRNAs | Expression | Target miRNAs | Target genes | Sample | Reference |
|---|---|---|---|---|---|
| circ19142 | Up | miR-7067-5p* | – | MC3T3-E1 cells | ( |
| circ5846 | Up | miR-7067-5p* | – | MC3T3-E1 cells | ( |
| circFgfr2 | Up | miR-133 | BMP6 | rDFCs | ( |
| mm9_circ_009056 | Up | miR-22-3p | BMP7 | MC3T3-E1 cells | ( |
| CDR1as | Up | miR-7 | GDF5 | hPDLSCs | ( |
| circMCM3AP | Down | miR-6881-3p | Smad6* | hASCs | ( |
| circPOMT1 | Down | miR-6881-3p | Smad6* | hASCs | ( |
| circ_AFF4 | Up | miR-135a-5p | FNDC5/Irisin/Smad1/5 | hBMSCs | ( |
| circSIPA1L1 | Up | miR-617 | Smad3 | hDPSCs | ( |
| circRNA124534 | Up | miR-496 | β-Catenin | hDPSCs | ( |
| circ_0024097 (circYAP1) | Up | miR-376b-3p | YAP1 | BMSCs, MC3T3-E1 cells | ( |
| CDR1as | Down | miR-7-5p | Wnt5b | SONFH-hBMSCs | ( |
| hsa_circ_0026827 | Up | miR-188-3p | RUNX1 | hDPSCs | ( |
| circ_0011269 | Up | miR-122 | RUNX2 | hBMSCs | ( |
| circRNA-23525 | Up | miR-30a-3p | RUNX2 | mASCs | ( |
| hsa_circ_0006215 | Up | miR-942-5p | RUNX2 | hBMSCs | ( |
| hsa_circ_0074834 | Up | miR-942-5p | ZEB1 | hBMSCs | ( |
| hsa_circ_33287 | Up | miR-214-3p | RUNX3 | hMSMSCs, HEK-293T | ( |
| circRNA_0006393 | Up | miR-145-5p | FOXO1 | hBMSCs | ( |
| has_circ_0001320 (circFOXP1) | Up | miR-33a-5p | FOXP1 | hASCs | ( |
*, target only predicted by bioinformatics. CircRNAs, circular RNAs; miRNAs, microRNAs; mRNAs, messenger RNAs. rDFCs, rat dental follicle cells; hPDLSCs, human periodontal ligament stem cells; hASCs, human adipose-derived stem cells; hDPSCs, human dental pulp stem cells; BMSCs, bone marrow stem cells; SONFH-hBMSCs, human bone marrow stem cells from steroid-induced osteonecrosis of the femoral head; mASCs, mouse adipose-derived stem cells; MSMSCs, maxillary sinus membrane stem cells; SCAPs, stem cells from apical papilla.
Smad-dependent BMPs/TGF-β
BMPs are a crucial member of the TGF-β superfamily. There are over 20 human encoded BMP ligands, among which BMP2, BMP4, BMP5, BMP6, BMP7, BMP9 (also referred to as GDF2), and BMP14 (also referred to as GDF5) have been reported to correlate well with osteogenesis (52-54). Research has shown that these BMPs play vital roles in skeletal development and postnatal bone remodeling by triggering canonical Smad-dependent or non-canonical p38 mitogen-activated protein kinase (MAPK) pathways (55,56). Smads are core mediators in both Smad-dependent BMP and TGF-β signaling. The eight encoded mammal Smads can be classified into the following three subtypes: (I) receptor-regulated Smads (R-Smads: Smad1, Smad2, Smad3, Smad5, and Smad8); (II) common-mediator Smads (Co-Smads: Smad4); and (III) inhibitory Smads (I-Smads: Smad6 and Smad7) (57,58). Once BMP ligands or TGF-β ligands bind to their receptors (tetramer comprised of 2 TGFβRI and 2 TGFβRII), Smad1/5/8 or Smad2/3, respectively, will be phosphorylated and compounded with Smad4. Next, the R-Smads-Co-Smad complex will be translocated into the nucleus to stimulate the expression of osteogenic genes. Additionally, the process mentioned above can be blocked by I-Smads (Smad6/7) (59,60). Figure 2 displays a sketch detailing the involvement of circRNAs in the Smad-dependent BMP/TGF-β pathways.
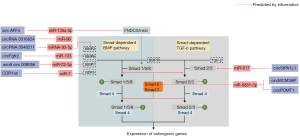
BMP2 is a well-known agent involved in the osteogenic process, and recombinant human BMP2 (rhBMP2) has been used in clinical therapy (61). Research has shown that osteogenic markers induced by human BMP2 are significantly upregulated in MC3T3-E1 cells (34). Further, RNA sequencing showed that 158 circRNAs were differentially expressed in a BMP2-treated group, which revealed the intricate connections between BMP2 and circRNAs in the skeletal system. Additionally, circ19142 and circ5846, which are 2 circRNAs upregulated in the BMP2 group, are predicted to be strongly associated with miR-7067-5p and the downstream osteogenic pathways (34).
Expression patterns of circRNAs have been detected in rat dental follicle cell (rDFC) osteogenic differentiation (35). Among the modulated circRNAs, circFgfr2 was observed to be upregulated via RNA high-throughput sequencing and quantitative real-time polymerase chain reaction (qRT-PCR). In situ hybridization showed that the expression trend of miR-133 was negatively correlated with those of circFgfr2 and BMP6 in rat dental follicle tissues from day 1 to 11 postpartum. In addition, the overexpression of circFgfr2 downregulated miR-133 enhanced the expression of BMP6 and osteogenic marker genes, indicating that circFgfr2/miR-133/BMP6 cascade is a potential regulatory mechanism underlying the process of osteogenesis. However, further investigations need to be conducted to determine their exact interplay and relationships in this type of circRNA-miRNA-mRNA network.
Calcitonin gene-related peptide (CGRP) has been reported to promote osteogenesis and has been applied in MC3T3-E1 cell osteogenic induction (36). The expression level of exon-derived mm9_circ_009056 was observed to be higher in a CGRP-supplemented osteogenic induction group than an osteogenic induction group, which suggests that mm9_circ_009056 is relevant to CGRP-induced osteogenesis. Additionally, miR-22-3p, which was downregulated in a CGRP-induced group, contained binding sites with both mm9_circ_009056 and BMP7 mRNA (36). Thus, BMP7 is a target gene regulated by an upstream competing endogenous RNA (ceRNA) network to orchestrate MC3T3-E1 cell osteogenesis (36).
The cerebellar degeneration-related protein 1 antisense (CDR1as) is a well-studied circRNA with sponge function toward miR-7, and the CDR1as-miR-7 interaction is crucial in the osteogenesis of hPDLSCs (37). Concerning the downstream target, the 3' untranslated region (3'-UTR) of GDF5 (also referred to as BMP14) was thought to contain binding sites with miR-7, and a luciferase reporter assay later confirmed this relationship. As GDF5 is a member of the BMP family, and could enhance osteogenic differentiation by activating the Smad-dependent BMP pathway and the MAPK pathway (55,56), the study further focused on whether CDR1as/miR-7/GDF5 cascade affected these two pathways. Interestingly, CDR1as and GDF5 knockdown or miR-7 overexpression severely impaired the phosphorylation of Smad1/5/8 and p38 MAPK, which suggests that they play important roles in osteogenesis-related pathways.
CircMCM3AP and circPOMT1 were reported to be downregulated during the osteogenesis of human adipose-derived stem cells (hASCs), and according to bioinformatic prediction results, miR-6881-3p is one of their co-targets (38). The expression of miR-6881-3p has been proven to be negatively related to circMCM3AP and circPOMT1, which suggests that these two circRNAs might sequester it. Smad6, an inhibitor of Smad-dependent BMP and TGF-β signaling, was predicted to be a possible direct target of miR-6881-3p.
It has been reported that circ_AFF4 sponged miR-135a-5p to regulate FNDC5/Irisin, the activation of which triggered the phosphorylation of Smad1/5 (39). As p-Smad1/5 are key mediators in the Smad-dependent BMP pathway, the overexpression of circ_AFF4 could positively regulate this pathway to promote osteogenesis. In another study, circSIPA1L1 was identified to stimulate the osteogenesis of hDPSCs by tethering miR-617, which directly targets Smad3 (40). As Smad3 is a pivotal component of R-Smads, the effects of the circSIPA1L1/miR-617/Smad3 circuit might be activated by a Smad-dependent TGF-β pathway.
Wnts
The best understood Wnt pathway is the canonical Wnt/β-Catenin pathway, which has been discussed in several reviews (62-64). Briefly, in the absence of Wnt, cytoplasmic β-Catenin proteins are phosphorylated by “destruction complexes” [compounds consisting of Axin, adenomatous polyposis coli (APC), glycogen synthase kinase-3β (GSK3β), and casein kinase 1α (CK1α)], resulting in the constant degradation of β-Catenin by the proteasome. Once bound with a transmembrane heterodimer composed of Frizzled (FZD) and LRP5/6, Wnt stabilizes β-Catenin by eliminating the degradation effect from the “destruction complexes”, and then translocating it into the nucleus to regulate the target genes (see Figure 3). Wnt signaling is involved in in biological processes, such as cell proliferation, cell fate determination, and postnatal tissue homeostasis (65). Notably, numerous studies have examined the close relationship between osteoblastic differentiation and the Wnt/β-Catenin pathway (66-68).

It has been revealed that a correlation exists between circRNAs and Wnt signaling during osteogenic differentiation. The sequencing of circRNAs and mRNAs were performed in several studies in which bioinformatic analyses showed the possible involvement of Wnt signaling (32,69). Additionally, circ19142 and circ5846, two circRNAs that are significantly upregulated in BMP-2-induced MC3T3-E1 cells, were predicted to target miR-7067-5p (34). Gene Ontology (GO) and PANTHER pathway analyses have shown that there is a strong relationship between the Wnt signaling pathway and the circRNAs-miRNA network, but these interactions need to be confirmed in further experiments. Another research group showed that the overexpression of circRNA124534 promoted hDPSC osteogenesis both in vitro and in vivo and regulated β-Catenin by directly sponging miR-496 (41). The suppression of miR-496 enhanced the osteogenic effect of circRNA124534; however, the improvement of miRNA expression produced opposite results. Another study (42) reported that the circ_0024097/miR-376b-3p/YAP1 cascade could trigger the Wnt/β-Catenin pathway, and further exploration showed that WIF-1 (a β-Catenin inhibitor) completely rescued the promoting effects of circ_0024097 on osteoblastic differentiation. As the expression levels of circRNA124534 and circ_0024097 are both positively related to that of β-Catenin, they may work as an osteogenic determinant via the canonical Wnt signaling cascade.
Concerning the non-canonical Wnt pathway, several Wnts (i.e., Wnt4, Wnt5a, and Wnt5b) have been reported to activate signaling cascades independent of β-Catenin, such as Wnt-Ca2+ and Wnt-atypical protein kinase C pathways (70). Additionally, those β-Catenin-independent Wnts may compete for co-receptors with β-Catenin-dependent Wnts on inhibiting the canonical Wnt pathway (71). One study focused on the steroid-induced osteonecrosis of the femoral head (SONFH), in which the adipogenic/osteogenic differentiation balance of the hBMSCs was disturbed (43). Under pathological conditions, highly abundant CDR1as upregulated Wnt5b to promote adipogenesis and inhibited osteogenesis by competitively harboring miR-7-5p (43). It was also observed that the knockdown of CDR1as and Wnt5b led to the accumulation of β-Catenin, confirming the negative correlation between Wnt5b and β-Catenin. Taken together, it is reasonable to hypothesize that the CDR1as/mi-7-5p/Wnt5b axis can activate the non-canonical Wnt pathway and competitively inhibit the Wnt/β-Catenin pathway, thus influencing the osteogenic differentiation of BMSCs.
In conclusion, the activation of canonical and non-canonical Wnt pathways may function oppositely to one other during the induction of osteogenesis, but the exact interaction and underlying mechanisms require further investigation.
RUNX
RUNX1, RUNX2, and RUNX3 are part of the mammalian RUNX transcription factor family involved in diverse biological activities, including skeletal development, hematopoiesis, and neurogenesis (72-74).
Hsa_circ_0026827 was reported to have osteogenic potential in in vitro experiments and in an in vivo heterotopic bone model (44). Additionally, a luciferase reporter assay confirmed the interaction between hsa_circ_0026827, miR-188-3p, and RUNX1 in the osteoblast differentiation process of hDPSCs. Hence, the acknowledged hematopoiesis regulator—RUNX1 (73), may also affect osteogenesis under the control of the ceRNA mechanism.
RUNX2, a key determinant of osteoblast differentiation and maturation (75), is broadly used as an osteogenic marker gene. Xu et al. showed that the expressions of circ_0011269 and RUNX2 were both upregulated, while the expression of miR-122 was downregulated during the osteogenic process (45). A subsequent study showed that circ_0011269 sequestered miR-122 to regulate the expression level of RUNX2. Similarly, the expression of circRNA-23525 was shown to increase with osteogenic induction gradually (46), and its alteration regulated osteogenic differentiation. MiR-30a-3p and RUNX2 were further confirmed to be their downstream agents (46). Hsa_circ_0006215, which increased significantly during the osteogenic differentiation of BMSCs and peaked on the 7th day of induction, was surprisingly found to enhance osteogenesis-angiogenesis coupling (47). A luciferase reporter assay found that hsa_circ_0006215 directly targeted miR-942-5p, and osteogenesis-angiogenesis effects occurred following the activation of RUNX2 and VEGF, which enabled bone tissue regeneration with neovascularization. Interestingly, miR-942-5p was discovered to be sponged by another upstream molecule, hsa_circ_0074834 (48), which suggests that the components in circRNAs-miRNAs-mRNA circuits interact with each other to form a complex network.
RUNX3 is also related to osteogenesis, albeit to a lesser extent. Notably, RUNX3-knockout mice developed severe congenital osteopenia, which was attributed to decreased osteoblast numbers and mineral deposition (76). Hsa_circ_33287 was screened out of microarray data as an osteogenic candidate (49). Follow-up tests confirmed the hypothesis and showed that hsa_circ_33287 targeted RUNX3 to stimulate the osteogenic differentiation of human maxillary sinus membrane stem cells (hMSMSCs) by serving as a miR-214-3p sponge.
FOXs
The FOX family includes a class of evolutionarily conserved transcriptional regulators. Their classification and contribution to bone metabolism were summarized in a recent review (77). Some FOX family members interact with genes, such as Runx2, Osx, and Col1, to promote the osteogenic differentiation of mesenchymal stem cells (78,79).
FOXO1 is a transcription factor that belongs to the forkhead box class O family proteins (FOXOs), which have been extensively detected in bone tissues (80,81). Ma et al. reviewed the regulatory function of FOXOs toward bone cells and found that FOXOs played critical roles in different types of bone physiological processes, such as bone development, bone remodeling, and energy metabolism (82). Notably, Wang et al. discovered that the expression level of circRNA_0006393 was decreased in glucocorticoid-induced osteoporosis (GIOP) patients (50). Additionally, the overexpression of circRNA_0006393 was shown to promote osteogenesis by sponging miR–145–5p and upregulating FOXO1. Thus, circRNA_0006393 can enhance osteogenesis as an upstream regulatory factor of FOXO1.
Another member of the FOX family involved in osteogenesis is forkhead box P1 (FOXP1), which also has the potential to affect the differentiation potency of mesenchymal stem cells (83). It is an interesting phenomenon that circRNAs regulate their parental genes and that such a relationship exists between FOXP1 and its circular counterpart (51). Highly presented circFOXP1 was reported to enhance FOXP1 expression during hASC osteogenic differentiation. This effect can be blocked by miR-33a-5p, which confirms that miR-33a-5p is a crucial mediator bridging circFOXP1 and FOXP1.
CircRNAs modulate osteoclasts and bone resorption
Osteoclasts are hematopoietic deriving multinucleated cells, which have bone resorption potential (84). Their bone and normal function presence play an important role in skeletal growth, modeling, and remodeling (84). Numerous regulatory agents have been discovered to control the differentiation of hematopoietic progenitors into mature osteoclasts (85). In the early stage of osteoclastogenesis, pivotal regulators include PU.1, which activates CSF-1R and microphthalmia transcription factor (MITF) (86,87). CSF-1 is vital to the survival and proliferation of osteoclast precursors, and it also facilitates the membrane RANK expression of precursor cells to ensure RANK-RANKL binding (87,88). After an interaction between RANK and RANKL, the differentiation of osteoclast precursors into preosteoclasts is initiated (84,85). Conversely, this process can be blocked by OPG (84,85). Fusion and polarization finally determine the mature osteoclasts, which function as bone-resorbing cells (89). This process is depicted in Figure 4. The roles of circRNAs in these processes are unknown; however, evidence suggests that diverse circRNAs are involved in osteoclastogenesis and bone resorption. It has been reported that circRNAs are differently expressed between osteoclast precursors and mature osteoclasts (90). Dou et al. induced RAW264.7 cells with RANKL and CSF-1 and then observed and verified osteoclastogenesis. They subsequently sequenced the expression profile of circRNAs and miRNAs, and found that 1 upregulated circRNA (circRNA_007873) and 2 downregulated circRNAs (circRNA_010763, circRNA_015622) all targeted the downstream miR-103 to facilitate the differentiation process (12). Thus, we seek to explore which circRNAs are involved in the critical pathways of osteoclastogenesis (CSF-1 and RANKL/OPG); the regulatory networks are listed in Table 2.

Table 2
| CircRNAs | Expression | Target miRNAs | Target genes | Sample | Reference |
|---|---|---|---|---|---|
| circRNA_007873 | Up | miR-103* | – | RAW264.7 cells | ( |
| circRNA_010763 | Down | miR-103* | – | RAW264.7 cells | ( |
| circRNA_015622 | Down | miR-103* | – | RAW264.7 cells | ( |
| circRNA_28313 | Up | miR-195a |
|
mBMM cells | ( |
| circRNA_009934 | Up | miR-5107 |
|
mBMM | ( |
| circ_0007059 | Down | miR-378 |
|
hBMSCs | ( |
*, target only predicted by bioinformatics. mBMM cells, mouse bone marrow monocyte/macrophage cells; circRNAs, circular RNAs; miRNAs, microRNAs; mRNAs, messenger RNAs; hBMSCs, human bone marrow stem cells.
CSF-1
CSF-1, also known as the macrophage colony-stimulating factor (M-CSF), is an important growth factor regulating the osteoclastogenesis process. It exists in soluble or membrane-bounded forms and can bind to its receptor CSF-1R (also known as c-Fms), which is normally present in low levels in hematopoietic stem cells (HSCs) but in higher levels in monocytes/macrophages and osteoclasts (94). The interplay of CSF-1 and CSF-1R initiates the survival and proliferation of osteoclast precursors (95). Differentially expressed circRNAs, miRNAs, and mRNAs were detected in a diosgenin (DIO; a kind of phytoestrogen)-treated group compared to a control group after ovariectomy (96). The upregulated rno_circRNA_016717, along with 7 differentially expressed key mRNAs (including CSF-1) and 60 miRNAs, were then selected to establish a circRNA-miRNA-mRNA co-expression network, which provided evidence of the potential function of circRNAs in CSF-1-mediated bone metabolism.
Bone marrow monocyte/macrophage (BMM) cells are the precursors of osteoclasts. They are often cultured in medium supplemented with CSF-1 and RANKL for osteoclastogenesis. In a recent study, over 10,000 differentially expressed circRNAs were discovered between differentiated and non-differentiated mouse BMM cells (91). Among the circRNAs mentioned above, circRNA_28313 was selected as a candidate for functional validation, as it was significantly upregulated during osteoclast differentiation. Its knockdown impaired tartrate-resistant acid phosphatase (TRAP)-positive multinuclear cell formation and the expression level of osteoclastogenic markers. In the downstream signals, miR-195a, a direct target of circRNA_28313, was found to bind with the 3'-UTR of CSF-1 and work as a CSF-1 suppressor. Thus, the circRNA_28313/ miR-195a/ CSF-1 feedback regulatory loop occurred during in vitro osteoclast differentiation. In an ovariectomized mouse model, micro-computed tomography (CT) scanning and histological analyses revealed that the bone loss of the circRNA_28313 depletion group was significantly less than that of the control group, which provided evidence of the osteoclast formation ability of circRNA_28313 in an in vivo context.
Another study reported that circASAP1 mediated tumor-associated-macrophage infiltration via the miR326/miR-532-5p-CSF-1 axis (97). In vitro experiments also verified that circASAP1 enhanced the proliferation and migration of macrophages, which could be offset by miR326/miR-532-5p overexpression or CSF-1 knockdown. Osteoclasts differ from bone tissue macrophages, which have the same hematopoietic progenitors of tumor-associated macrophages. Thus, it is reasonable to hypothesize that circASAP1 may similarly work as an upstream regulator toward CSF-1 via miR-326 and miR-532-5p during the process of osteoclastogenesis. However, this hypothesis requires further investigation.
RANKL/OPG
RANKL (also known as OPGL or TRANCE) is expressed by some immune cells (e.g., monocytes, T and B cells, and dendritic cells) and osteoclastogenesis-supporting cells (e.g., osteoblasts, osteocytes, and chondrocytes) (87,98). Once RANKL binds to the RANK on osteoclast precursors’ surface, differentiation signals are transduced by recruiting molecules of TNF receptor-associated factor (TRAF) family, among which TRAF6 has been confirmed to be a main transduction factor, and facilitate the activation of nuclear factor kappa B (NF-κB) and MAPKs (98). OPG, a soluble decoy ligand of RANKL, has a bone-protecting potential due to its ability to neutralize RANKL (98).
Research has shown that the differentiation process of osteoclasts is regulated by the relative expression level of RANKL/OPG, and diverse agents cross-talking with these two ligands could affect this process (99). For example, circRNAs related to osteoclast differentiation were selected to construct a circRNA-miRNA-mRNA network (90). Among the modulated mRNAs, NF-κB1, and TRAF6, which are indispensable mediators of the RANK/RANKL pathway, were predicted to be targeted by upstream miRNAs and circRNAs. Further well-designed experiments need to be conducted to validate this interaction; however, these results indicate a possible link between circRNAs and RANKL-related mechanisms in osteoclastogenesis.
It has been suggested that CircRNA_009934, which is highly expressed in mature osteoclasts, directly sequesters miR-5107 via a luciferase reporter assay (92). As for the specific downstream target gene, research has shown that miR-5107 mimics significantly suppress mRNA and protein expression of TRAF6 in mBMM cells, and the knockdown of the miRNA rescued TRAF6 inhibition from circRNA depletion.
CircRNAs in bone-remodeling disorders
During bone-remodeling procedures, the physiological structure and function of the bone can only be perfectly maintained when the metabolism activities of osteoblasts and osteoclasts are fine-tuned to keep in balance. If this balance is disrupted, a series of bone-related diseases occur, including osteoporosis, Paget’s disease, and osteopetrosis (7). Epigenetic regulators play an important role in such pathological processes (100-102). Table 3 summarizes the relevant circRNAs.
Table 3
| CircRNAs | Expression | Sample | Reference |
|---|---|---|---|
| hsa_circ_0001275 | Up | blood samples from PMOP patients | ( |
| circRNA_0016624 | Down | blood samples from PMOP patients | ( |
| circRNA_0048211 | Down | bone samples from PMOP patients | ( |
| circ_0007059 | Down | samples from PMOP patients | ( |
| hsa_circ_0006393 | Down | bone samples from GIOP patients | ( |
| circUSP45 | Up | bone samples from SONFH patients | ( |
| circRNA_25487 | Up | blood samples from TONFH patients | ( |
CircRNAs, circular RNAs; PMOP, postmenopausal osteoporosis; GIOP, glucocorticoid-induced osteoporosis; SONFH, steroid-induced osteonecrosis of the femoral head; TONFH, trauma-induced osteonecrosis of femoral head.
Postmenopausal osteoporosis (PMOP)
PMOP is a kind of primary osteoporosis directly caused by estrogen deficiency. Under this pathological condition, bone resorption exceeds bone formation, which increases the risk of bone fracture in older women (7). Due to rapid developments in next-generation sequencing technology, correlation between PMOP and circRNAs has been verified in several studies. A receiver operating characteristic (ROC) analysis of the peripheral blood mononuclear cells (PBMCs) of PMOP patients showed that hsa_circ_0001275 has a significant diagnostic value in PMOP (103). Additionally, the RNA sequencing of BMSCs collected from mice who underwent an ovariectomy (a surgery usually used to simulate PMOP) showed the circRNA-associated ceRNA network in the PMOP mouse model (101). These results suggest that circRNAs have a regulatory effect upon PMOP both in vivo and in vitro.
In studies focusing on the mechanisms of PMOP, blood and bone samples were collected for circRNA expression detection (93,104,105). circRNA_0016624 and circRNA_0048211 were both reported to be downregulated in PMOP patient specimens. CircRNA_0016624 and circRNA_0048211 were observed to be abundantly expressed during hBMSC osteogenic differentiation, and their effects upon osteogenesis were mediated by miR-98 and miRNA-93-5p, respectively (104,105). Intriguingly, BMP2 was the co-target of these 2 circRNA-miRNA circuits, which shows the indispensable role of BMP2 in the circRNA regulatory network during the occurrence and progression of PMOP. As BMP2 is closely related to the osteogenic process, these results suggest that aberrantly expressed circRNAs might impair the differentiation and function of osteoblasts, thus facilitating the occurrence and progression of PMOP.
The deterioration of bone architecture in PMOP is also affected by osteoclast-regulated bone-resorbing. Liu et al. observed that circRNA_0007059 was involved in the RANKL-induced osteoclastogenesis process (93), during which its expression level gradually decreased. Concerning circ_0007059 overexpression, nuclear factor of activated T cells, cytoplasmic 1 (NFATc1) and TRAF6, which are key effectors of RANKL-induced signaling, were lowly expressed. Additionally, a dual-luciferase reporter assay validated the downstream effects of miR-378 and BMP2, suggesting that this interaction could retard RANKL-induced osteoclast differentiation.
Glucocorticoid-induced osteoporosis
GIOP, which is another type of osteoporosis, is characterized as a secondary disease initiated by the administration of glucocorticoids (108). However, its underlying mechanisms, especially those that involve circRNAs, remain unclear. Wang et al. (50) used samples from male patients with GIOP or traumatic fracture to avoid the effects of estrogen. Hsa_circ_0006393, which is derived from exons of the hypoxia-inducible factor 1α gene, was more downregulated in the GIOP group than the fracture group. In vivo, plasmid overexpressing hsa_circ_0006393 or dexamethasone were injected intramedullary into the femur of mice. The groups injected with dexamethasone and the hsa_circ_0006393 overexpressing vector had higher bone mineral density than those that received dexamethasone injections only. Thus, hsa_circ_0006393 appears to play a role in saving bone mass loss from glucocorticoids. However, more GIOP animal models need to be established to support this conclusion.
Osteonecrosis of the femoral head
Osteonecrosis, which is also known as avascular necrosis, is associated with traumatic and non-traumatic conditions (109). Different etiological factors cause hypoperfusion of the bone, which results in ischemia and the subsequent disturbance of bone cells (i.e., osteocytes, osteoblasts, and macrophage populations) and the microenvironment (110). Thus, the impairment of physiological bone-remodeling processes and the collapse of subchondral bone occur. Due to the anatomy of the vascular supply, the femoral head is especially prone to osteonecrosis (111). Recently, it was discovered that the expression of circRNAs was related to the occurrence of osteonecrosis of the femoral head (ONFH) (112). Jiao et al. (112) acquired subchondral bone from both ONFH patients and fracture patients. The transcriptomes of these two groups showed that 44 circRNAs were upregulated and 30 circRNAs were downregulated in the ONFH sample.
The application of corticosteroids contributes to the pathogenesis of SONFH, which is refractory and severely affects patients’ quality of life. Pharmacologic or physical treatment and surgical techniques have been introduced to treat SONFH (113), but problems, such as the undesirable regeneration of physiological bone structures and functions, remain. There is extensive evidence of the osteoinductive effects of CircRNAs. Thus, circRNAs represent promising molecules for bone regenerative therapies. Hao et al. constructed SONFH models in vitro and in vivo (114). CircPVT1, which has been reported in several tumors, was detected to be downregulated in glucocorticoid-treated hBMSCs. The overexpression of circPVT1 was found to promote osteogenic differentiation in vitro, improve bone tissue morphology, and attenuate SONFH in a rat model by targeting downstream miR-21-5p and Smad7. Other research has demonstrated the adverse effects of circRNAs in SONFH (106). Using high-throughput sequencing, circUSP45 was selected as key upregulated circRNA in specimens from SONFH patients, which indicates that it may be involved in the progression of SONFH. Subsequent experiments showed that si-circUSP45 saved the femoral head from dexamethasone-induced bone tissue deteriorates. The Phosphatase and TENsin homolog (PTEN), which is a crucial gene regulating cell proliferation, was discovered to be epigenetically regulated by circUSP45-miRNA-127-5p, which suggests the circUSP45–miRNA-127-5p–PTEN cascade is a possible pathway in the pathological process of SONFH.
Femoral neck fractures or other kinds of trauma that cause vascular compromise to the femoral head can be attributed to the occurrence of trauma-induced osteonecrosis of the femoral head (TONFH) (109). CircRNAs have been reported to be aberrantly expressed in patients with TONFH compared to those who have healed from femoral neck fractures (107). Among these circRNAs, circRNA_25487 was significantly upregulated in the peripheral blood of TONFH patients. Further, miRNA-134-3p was shown to interact with circRNA_25487 and downstream p21 mRNA, which indicates that it may be a potential target for the treatment of TONFH.
Osteopetrosis
Osteopetrosis, also called marble bone disease, refers to a series of inherited diseases (classified as autosomal dominant, autosomal recessive, and X-linked based on inheritance) that result from osteoclast differentiation and function impairment (115). The dysfunction of osteoclasts causes the imbalance of bone remodeling and thus, increases bone density.
Several studies have found a correlation between miRNAs and osteopetrosis. In vitro, the silencing of DiGeorge Critical Region 8 (DGCR8), Dicer, or Mammalian Argonaute 2 (Ago2), which are pivotal factors during the miRNA generating and homeostasis process, were shown to impair osteoclastogenesis and bone resorption (116) significantly. Such effects were further validated in a transgenic mouse model in which CD11b(+)-cre/Dicer-null mice showed mild osteopetrosis. Additionally, via deep sequencing and iTRAQ quantitative proteomics, 123 differently expressed miRNAs and 173 differently expressed proteins were detected in the PBMCs of osteopetrosis patients and healthy donors (102). Arf1 was further predicted and validated to be a target gene of has-miR-320a. Thus, research suggests that miRNAs play a role in the pathology process of osteopetrosis. Given the well-known sponge function of circRNAs toward miRNAs, the effects of miRNAs present in osteopetrosis may be affected by a negative feedback loop of their upstream circRNAs.
Conclusion and perspective
Collectively, many circRNAs have been shown to promote osteogenesis and facilitate osteoclast differentiation via diverse mechanisms, and thus modulate the process of bone homeostasis. The imbalance or impairment of these two parts causes diseases, such as osteoporosis, and osteonecrosis of the femoral head, which are also closely correlated to the aberrant presence of circRNAs. Current evidence provides us with promising diagnosis and treatment methods for some bone homeostasis disorders.
However, deficiencies remain to be addressed. A great number of studies have detected circRNA expression during the osteogenic differentiation of different kinds of cells. Interestingly, the results showed different expression profiles (29-33); however, this might be related to the different kinds and species of the cells. Additionally, under different conditions, the tissue sources may also affect the function of circRNAs. For example, the upregulation of CDR1as was found to have opposite functions in osteogenesis in 2 different studies (37,43). This may be because these 2 studies isolated cells from tissues in physiological and pathological conditions, respectively. Other factors, such as the culture medium and time of induction, could also have affected the results. More evidence needs to be gathered to interpret this phenomenon and explore how to apply the results of studies from the laboratory to the clinic. To date, studies have mostly focused on circRNAs’ promotion of osteogenesis and very few researches have examined the roles of circRNAs in osteoclastogenesis, which is of equal importance. Currently, research has focused on investigating the sponge function of circRNAs in the bone-formation and bone-absorption processes. Thus far, the other completely different but equally important functions have rarely been mentioned. Well-designed in-vivo experiments are required to further explore the effects of circRNAs in the living body context to realize their early clinical application in disease diagnosis and treatment.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (No. 81771048 to ZZ, No. 81900981 to XC); China Postdoctoral Science Foundation (No. 2019M663530 to XC); Sichuan Science and Technology Program (No. 2020YFS0170 to ZZ, No. 2021YJ0149 to XC); Research and Development Foundation of West China Hospital of Stomatology (No. RD-02-201904 to XC); and Research Funding for Talents Developing, West China Hospital of Stomatology Sichuan University (No. RCDWJS2020-18 to XC).
Footnote
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-2114
Peer Review File: Available at https://dx.doi.org/10.21037/atm-21-2114
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-2114). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Berendsen AD, Olsen BR. Bone development. Bone 2015;80:14-8. [Crossref] [PubMed]
- Seeman E, Delmas PD. Bone quality--the material and structural basis of bone strength and fragility. N Engl J Med 2006;354:2250-61. [Crossref] [PubMed]
- Harada S, Rodan GA. Control of osteoblast function and regulation of bone mass. Nature 2003;423:349-55. [Crossref] [PubMed]
- Kim JM, Lin C, Stavre Z, et al. Osteoblast-Osteoclast Communication and Bone Homeostasis. Cells 2020;9:2073. [Crossref] [PubMed]
- Langdahl B, Ferrari S, Dempster DW. Bone modeling and remodeling: potential as therapeutic targets for the treatment of osteoporosis. Ther Adv Musculoskelet Dis 2016;8:225-35. [Crossref] [PubMed]
- Siddiqui JA, Partridge NC. Physiological Bone Remodeling: Systemic Regulation and Growth Factor Involvement. Physiology (Bethesda) 2016;31:233-45. [Crossref] [PubMed]
- Feng X, McDonald JM. Disorders of bone remodeling. Annu Rev Pathol 2011;6:121-45. [Crossref] [PubMed]
- Wang L, Tong X, Zhou Z, et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-β-induced epithelial-mesenchymal transition and metastasis by controlling TIF1γ in non-small cell lung cancer. Mol Cancer 2018;17:140. [Crossref] [PubMed]
- Guerra BS, Lima J, Araujo B, et al. Biogenesis of circular RNAs and their role in cellular and molecular phenotypes of neurological disorders. Semin Cell Dev Biol 2021;114:1-10. [Crossref] [PubMed]
- Tang Y, Bao J, Hu J, et al. Circular RNA in cardiovascular disease: Expression, mechanisms and clinical prospects. J Cell Mol Med 2021;25:1817-24. [Crossref] [PubMed]
- Zhang M, Jia L, Zheng Y. circRNA Expression Profiles in Human Bone Marrow Stem Cells Undergoing Osteoblast Differentiation. Stem Cell Rev Rep 2019;15:126-38. [Crossref] [PubMed]
- Dou C, Cao Z, Yang B, et al. Changing expression profiles of lncRNAs, mRNAs, circRNAs and miRNAs during osteoclastogenesis. Sci Rep 2016;6:21499. [Crossref] [PubMed]
- Starke S, Jost I, Rossbach O, et al. Exon circularization requires canonical splice signals. Cell Rep 2015;10:103-11. [Crossref] [PubMed]
- Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol 2020;21:475-90. [Crossref] [PubMed]
- Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 2016;17:205-11. [Crossref] [PubMed]
- Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 2019;20:675-91. [Crossref] [PubMed]
- Shi X, Wang B, Feng X, et al. circRNAs and Exosomes: A Mysterious Frontier for Human Cancer. Mol Ther Nucleic Acids 2020;19:384-92. [Crossref] [PubMed]
- Bach DH, Lee SK, Sood AK. Circular RNAs in Cancer. Mol Ther Nucleic Acids 2019;16:118-29. [Crossref] [PubMed]
- Guo JU, Agarwal V, Guo H, et al. Expanded identification and characterization of mammalian circular RNAs. Genome Biol 2014;15:409. [Crossref] [PubMed]
- Sanger HL, Klotz G, Riesner D, et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A 1976;73:3852-6. [Crossref] [PubMed]
- Nigro JM, Cho KR, Fearon ER, et al. Scrambled exons. Cell 1991;64:607-13. [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Mol Cell 2017;66:9-21.e7. [Crossref] [PubMed]
- Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 1995;268:415-7. [Crossref] [PubMed]
- Zhang M, Zhao K, Xu X, et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun 2018;9:4475. [Crossref] [PubMed]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 2014;56:55-66. [Crossref] [PubMed]
- Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015;22:256-64. [Crossref] [PubMed]
- Kang Y, Guo S, Sun Q, et al. Differential circular RNA expression profiling during osteogenic differentiation in human adipose-derived stem cells. Epigenomics 2020;12:289-302. [Crossref] [PubMed]
- Long T, Guo Z, Han L, et al. Differential Expression Profiles of Circular RNAs During Osteogenic Differentiation of Mouse Adipose-Derived Stromal Cells. Calcif Tissue Int 2018;103:338-52. [Crossref] [PubMed]
- Li Z, Li N, Ge X, et al. Differential circular RNA expression profiling during osteogenic differentiation of stem cells from apical papilla. Epigenomics 2019;11:1057-73. [Crossref] [PubMed]
- Gu X, Li M, Jin Y, et al. Identification and integrated analysis of differentially expressed lncRNAs and circRNAs reveal the potential ceRNA networks during PDLSC osteogenic differentiation. BMC Genet 2017;18:100. [Crossref] [PubMed]
- Chen M, Yang Y, Zeng J, et al. circRNA Expression Profile in Dental Pulp Stem Cells during Odontogenic Differentiation. Stem Cells Int 2020;2020:5405931 [Crossref] [PubMed]
- Qian DY, Yan GB, Bai B, et al. Differential circRNA expression profiles during the BMP2-induced osteogenic differentiation of MC3T3-E1 cells. Biomed Pharmacother 2017;90:492-9. [Crossref] [PubMed]
- Du Y, Li J, Hou Y, et al. Alteration of circular RNA expression in rat dental follicle cells during osteogenic differentiation. J Cell Biochem 2019;120:13289-301. [Crossref] [PubMed]
- Wu C, Zheng Z, Ren W, et al. Mm9_circ_009056 enhances osteogenesis by targeting BMP7 via CGRP-mediated miR-22-3p. Biochem Biophys Res Commun 2018;501:199-205. [Crossref] [PubMed]
- Li X, Zheng Y, Zheng Y, et al. Circular RNA CDR1as regulates osteoblastic differentiation of periodontal ligament stem cells via the miR-7/GDF5/SMAD and p38 MAPK signaling pathway. Stem Cell Res Ther 2018;9:232. [Crossref] [PubMed]
- Huang XQ, Cen X, Sun WT, et al. CircPOMT1 and circMCM3AP inhibit osteogenic differentiation of human adipose-derived stem cells by targeting miR-6881-3p. Am J Transl Res 2019;11:4776-88. [PubMed]
- Liu C, Liu AS, Zhong D, et al. Circular RNA AFF4 modulates osteogenic differentiation in BM-MSCs by activating SMAD1/5 pathway through miR-135a-5p/FNDC5/Irisin axis. Cell Death Dis 2021;12:631. [Crossref] [PubMed]
- Ge X, Li Z, Zhou Z, et al. Circular RNA SIPA1L1 promotes osteogenesis via regulating the miR-617/Smad3 axis in dental pulp stem cells. Stem Cell Res Ther 2020;11:364. [Crossref] [PubMed]
- Ji F, Pan J, Shen Z, et al. The Circular RNA circRNA124534 Promotes Osteogenic Differentiation of Human Dental Pulp Stem Cells Through Modulation of the miR-496/β-Catenin Pathway. Front Cell Dev Biol 2020;8:230. [Crossref] [PubMed]
- Huang Y, Xiao D, Huang S, et al. Circular RNA YAP1 attenuates osteoporosis through up-regulation of YAP1 and activation of Wnt/β-catenin pathway. Biomed Pharmacother 2020;129:110365 [Crossref] [PubMed]
- Chen G, Wang Q, Li Z, et al. Circular RNA CDR1as promotes adipogenic and suppresses osteogenic differentiation of BMSCs in steroid-induced osteonecrosis of the femoral head. Bone 2020;133:115258 [Crossref] [PubMed]
- Ji F, Zhu L, Pan J, et al. hsa_circ_0026827 Promotes Osteoblast Differentiation of Human Dental Pulp Stem Cells Through the Beclin1 and RUNX1 Signaling Pathways by Sponging miR-188-3p. Front Cell Dev Biol 2020;8:470. [Crossref] [PubMed]
- Xu X, Chen Y, Tan B, et al. Circular RNA circ_0011269 sponges miR-122 to regulate RUNX2 expression and promotes osteoporosis progression. J Cell Biochem 2020; Epub ahead of print. [Crossref] [PubMed]
- Guo Z, Zhao L, Ji S, et al. CircRNA-23525 regulates osteogenic differentiation of adipose-derived mesenchymal stem cells via miR-30a-3p. Cell Tissue Res 2021;383:795-807. [Crossref] [PubMed]
- Ji H, Cui X, Yang Y, et al. CircRNA hsa_circ_0006215 promotes osteogenic differentiation of BMSCs and enhances osteogenesis-angiogenesis coupling by competitively binding to miR-942-5p and regulating RUNX2 and VEGF. Aging (Albany NY) 2021;13:10275-88. [Crossref] [PubMed]
- Ouyang Z, Tan T, Zhang X, et al. CircRNA hsa_circ_0074834 promotes the osteogenesis-angiogenesis coupling process in bone mesenchymal stem cells (BMSCs) by acting as a ceRNA for miR-942-5p. Cell Death Dis 2019;10:932. [Crossref] [PubMed]
- Peng W, Zhu S, Chen J, et al. Hsa_circRNA_33287 promotes the osteogenic differentiation of maxillary sinus membrane stem cells via miR-214-3p/Runx3. Biomed Pharmacother 2019;109:1709-17. [Crossref] [PubMed]
- Wang XB, Li PB, Guo SF, et al. circRNA_0006393 promotes osteogenesis in glucocorticoid-induced osteoporosis by sponging miR-145-5p and upregulating FOXO1. Mol Med Rep 2019;20:2851-8. [Crossref] [PubMed]
- Shen W, Sun B, Zhou C, et al. CircFOXP1/FOXP1 promotes osteogenic differentiation in adipose-derived mesenchymal stem cells and bone regeneration in osteoporosis via miR-33a-5p. J Cell Mol Med 2020;24:12513-24. [Crossref] [PubMed]
- Wang EA. Bone morphogenetic proteins (BMPs): therapeutic potential in healing bony defects. Trends Biotechnol 1993;11:379-83. [Crossref] [PubMed]
- Samartzis D, Khanna N, Shen FH, et al. Update on bone morphogenetic proteins and their application in spine surgery. J Am Coll Surg 2005;200:236-48. [Crossref] [PubMed]
- Rahman MS, Akhtar N, Jamil HM, et al. TGF-β/BMP signaling and other molecular events: regulation of osteoblastogenesis and bone formation. Bone Res 2015;3:15005. [Crossref] [PubMed]
- Wu M, Chen G, Li YP. TGF-β and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res 2016;4:16009. [Crossref] [PubMed]
- Lowery JW, Rosen V. The BMP Pathway and Its Inhibitors in the Skeleton. Physiol Rev 2018;98:2431-52. [Crossref] [PubMed]
- ten Dijke P, Hill CS. New insights into TGF-beta-Smad signalling. Trends Biochem Sci 2004;29:265-73. [Crossref] [PubMed]
- Fennen M, Pap T, Dankbar B. Smad-dependent mechanisms of inflammatory bone destruction. Arthritis Res Ther 2016;18:279. [Crossref] [PubMed]
- Massagué J. TGFβ signalling in context. Nat Rev Mol Cell Biol 2012;13:616-30. [Crossref] [PubMed]
- Wrana JL. Signaling by the TGFβ superfamily. Cold Spring Harb Perspect Biol 2013;5:a011197 [Crossref] [PubMed]
- Lo KW, Ulery BD, Ashe KM, et al. Studies of bone morphogenetic protein-based surgical repair. Adv Drug Deliv Rev 2012;64:1277-91. [Crossref] [PubMed]
- Nusse R, Clevers H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 2017;169:985-99. [Crossref] [PubMed]
- MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell 2009;17:9-26. [Crossref] [PubMed]
- Lories RJ, Corr M, Lane NE. To Wnt or not to Wnt: the bone and joint health dilemma. Nat Rev Rheumatol 2013;9:328-39. [Crossref] [PubMed]
- Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol 2004;20:781-810. [Crossref] [PubMed]
- Zhao X, Xie L, Wang Z, et al. ZBP1 (DAI/DLM-1) promotes osteogenic differentiation while inhibiting adipogenic differentiation in mesenchymal stem cells through a positive feedback loop of Wnt/β-catenin signaling. Bone Res 2020;8:12. [Crossref]
- Maeda K, Kobayashi Y, Koide M, et al. The Regulation of Bone Metabolism and Disorders by Wnt Signaling. Int J Mol Sci 2019;20:5525. [Crossref] [PubMed]
- Duan P, Bonewald LF. The role of the wnt/β-catenin signaling pathway in formation and maintenance of bone and teeth. Int J Biochem Cell Biol 2016;77:23-9. [Crossref] [PubMed]
- Zhong W, Li X, Pathak JL, et al. Dicalcium silicate microparticles modulate the differential expression of circRNAs and mRNAs in BMSCs and promote osteogenesis via circ_1983-miR-6931-Gas7 interaction. Biomater Sci 2020;8:3664-77. [Crossref] [PubMed]
- Grumolato L, Liu G, Mong P, et al. Canonical and noncanonical Wnts use a common mechanism to activate completely unrelated coreceptors. Genes Dev 2010;24:2517-30. [Crossref] [PubMed]
- Kessenbrock K, Dijkgraaf GJ, Lawson DA, et al. A role for matrix metalloproteinases in regulating mammary stem cell function via the Wnt signaling pathway. Cell Stem Cell 2013;13:300-13. [Crossref] [PubMed]
- Mevel R, Draper JE, Lie-A-Ling M, et al. RUNX transcription factors: orchestrators of development. Development 2019;146:dev148296 [Crossref] [PubMed]
- de Bruijn M, Dzierzak E. Runx transcription factors in the development and function of the definitive hematopoietic system. Blood 2017;129:2061-9. [Crossref] [PubMed]
- Inoue K, Ozaki S, Shiga T, et al. Runx3 controls the axonal projection of proprioceptive dorsal root ganglion neurons. Nat Neurosci 2002;5:946-54. [Crossref] [PubMed]
- Komori T. Runx2, an inducer of osteoblast and chondrocyte differentiation. Histochem Cell Biol 2018;149:313-23. [Crossref] [PubMed]
- Bauer O, Sharir A, Kimura A, et al. Loss of osteoblast Runx3 produces severe congenital osteopenia. Mol Cell Biol 2015;35:1097-109. [Crossref] [PubMed]
- Huang J, Shen G, Ren H, et al. Role of forkhead box gene family in bone metabolism. J Cell Physiol 2020;235:1986-94. [Crossref] [PubMed]
- Teixeira CC, Liu Y, Thant LM, et al. Foxo1, a novel regulator of osteoblast differentiation and skeletogenesis. J Biol Chem 2010;285:31055-65. [Crossref] [PubMed]
- You W, Fan L, Duan D, et al. Foxc2 over-expression in bone marrow mesenchymal stem cells stimulates osteogenic differentiation and inhibits adipogenic differentiation. Mol Cell Biochem 2014;386:125-34. [Crossref] [PubMed]
- Chen D, Gong Y, Xu L, et al. Bidirectional regulation of osteogenic differentiation by the FOXO subfamily of Forkhead transcription factors in mammalian MSCs. Cell Prolif 2019;52:e12540 [Crossref] [PubMed]
- Carter ME, Brunet A. FOXO transcription factors. Curr Biol 2007;17:R113-4. [Crossref] [PubMed]
- Ma X, Su P, Yin C, et al. The Roles of FoxO Transcription Factors in Regulation of Bone Cells Function. Int J Mol Sci 2020;21:692. [Crossref] [PubMed]
- Li H, Liu P, Xu S, et al. FOXP1 controls mesenchymal stem cell commitment and senescence during skeletal aging. J Clin Invest 2017;127:1241-53. [Crossref] [PubMed]
- Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature 2003;423:337-42. [Crossref] [PubMed]
- Teitelbaum SL, Ross FP. Genetic regulation of osteoclast development and function. Nat Rev Genet 2003;4:638-49. [Crossref] [PubMed]
- Boyce BF. Advances in the regulation of osteoclasts and osteoclast functions. J Dent Res 2013;92:860-7. [Crossref] [PubMed]
- Takayanagi H. Osteoimmunology: shared mechanisms and crosstalk between the immune and bone systems. Nat Rev Immunol 2007;7:292-304. [Crossref] [PubMed]
- Park JH, Lee NK, Lee SY. Current Understanding of RANK Signaling in Osteoclast Differentiation and Maturation. Mol Cells 2017;40:706-13. [PubMed]
- Weitzmann MN, Ofotokun I. Physiological and pathophysiological bone turnover - role of the immune system. Nat Rev Endocrinol 2016;12:518-32. [Crossref] [PubMed]
- Lin J, Ma S, Zhu C, et al. Circular RNA atlas in osteoclast differentiation with and without alendronate treatment. J Orthop Surg Res 2020;15:240. [Crossref] [PubMed]
- Chen X, Ouyang Z, Shen Y, et al. CircRNA_28313/miR-195a/CSF1 axis modulates osteoclast differentiation to affect OVX-induced bone absorption in mice. RNA Biol 2019;16:1249-62. [Crossref] [PubMed]
- Miao F, Yin BH, Zhang X, et al. CircRNA_009934 induces osteoclast bone resorption via silencing miR-5107. Eur Rev Med Pharmacol Sci 2020;24:7580-8. [PubMed]
- Liu S, Wang C, Bai J, et al. Involvement of circRNA_0007059 in the regulation of postmenopausal osteoporosis by promoting the microRNA-378/BMP-2 axis. Cell Biol Int 2021;45:447-55. [Crossref] [PubMed]
- Stanley ER, Chitu V. CSF-1 receptor signaling in myeloid cells. Cold Spring Harb Perspect Biol 2014;6:a021857 [Crossref] [PubMed]
- Kumari A, Silakari O, Singh RK. Recent advances in colony stimulating factor-1 receptor/c-FMS as an emerging target for various therapeutic implications. Biomed Pharmacother 2018;103:662-79. [Crossref] [PubMed]
- Zhang Z, Yue L, Wang Y, et al. A circRNA-miRNA-mRNA network plays a role in the protective effect of diosgenin on alveolar bone loss in ovariectomized rats. BMC Complement Med Ther 2020;20:220. [Crossref] [PubMed]
- Hu ZQ, Zhou SL, Li J, et al. Circular RNA Sequencing Identifies CircASAP1 as a Key Regulator in Hepatocellular Carcinoma Metastasis. Hepatology 2020;72:906-22. [Crossref] [PubMed]
- Nakashima T, Hayashi M, Takayanagi H. New insights into osteoclastogenic signaling mechanisms. Trends Endocrinol Metab 2012;23:582-90. [Crossref] [PubMed]
- Martin TJ, Sims NA. RANKL/OPG; Critical role in bone physiology. Rev Endocr Metab Disord 2015;16:131-9. [Crossref] [PubMed]
- Jin D, Wu X, Yu H, et al. Systematic analysis of lncRNAs, mRNAs, circRNAs and miRNAs in patients with postmenopausal osteoporosis. Am J Transl Res 2018;10:1498-510. [PubMed]
- Wang H, Zhou K, Xiao F, et al. Identification of circRNA-associated ceRNA network in BMSCs of OVX models for postmenopausal osteoporosis. Sci Rep 2020;10:10896. [Crossref] [PubMed]
- Ou M, Zhang X, Dai Y, et al. Identification of potential microRNA-target pairs associated with osteopetrosis by deep sequencing, iTRAQ proteomics and bioinformatics. Eur J Hum Genet 2014;22:625-32. [Crossref] [PubMed]
- Zhao K, Zhao Q, Guo Z, et al. Hsa_Circ_0001275: A Potential Novel Diagnostic Biomarker for Postmenopausal Osteoporosis. Cell Physiol Biochem 2018;46:2508-16. [Crossref] [PubMed]
- Yu L, Liu Y. circRNA_0016624 could sponge miR-98 to regulate BMP2 expression in postmenopausal osteoporosis. Biochem Biophys Res Commun 2019;516:546-50. [Crossref] [PubMed]
- Qiao L, Li CG, Liu D. CircRNA_0048211 protects postmenopausal osteoporosis through targeting miRNA-93-5p to regulate BMP2. Eur Rev Med Pharmacol Sci 2020;24:3459-66. [PubMed]
- Kuang MJ, Xing F, Wang D, et al. CircUSP45 inhibited osteogenesis in glucocorticoid-induced osteonecrosis of femoral head by sponging miR-127-5p through PTEN/AKT signal pathway: Experimental studies. Biochem Biophys Res Commun 2019;509:255-61. [Crossref] [PubMed]
- Zhang Y, Jia S, Wei Q, et al. CircRNA_25487 inhibits bone repair in trauma-induced osteonecrosis of femoral head by sponging miR-134-3p through p21. Regen Ther 2021;16:23-31. [Crossref] [PubMed]
- O'Brien CA, Jia D, Plotkin LI, et al. Glucocorticoids act directly on osteoblasts and osteocytes to induce their apoptosis and reduce bone formation and strength. Endocrinology 2004;145:1835-41. [Crossref] [PubMed]
- Petek D, Hannouche D, Suva D. Osteonecrosis of the femoral head: pathophysiology and current concepts of treatment. EFORT Open Rev 2019;4:85-97. [Crossref] [PubMed]
- Elgaz S, Bonig H, Bader P. Mesenchymal stromal cells for osteonecrosis. J Transl Med 2020;18:399. [Crossref] [PubMed]
- Chang C, Greenspan A, Gershwin ME. The pathogenesis, diagnosis and clinical manifestations of steroid-induced osteonecrosis. J Autoimmun 2020;110:102460 [Crossref] [PubMed]
- Jiao M, Tian R, Liu G, et al. Circular RNA and Messenger RNA Expression Profile and Competing Endogenous RNA Network in Subchondral Bone in Osteonecrosis of the Femoral Head. DNA Cell Biol 2021;40:61-9. [Crossref] [PubMed]
- Zhao D, Zhang F, Wang B, et al. Guidelines for clinical diagnosis and treatment of osteonecrosis of the femoral head in adults (2019 version). J Orthop Translat 2020;21:100-10. [Crossref] [PubMed]
- Hao Y, Lu C, Zhang B, et al. CircPVT1 up-regulation attenuates steroid-induced osteonecrosis of the femoral head through regulating miR-21-5p-mediated Smad7/TGFβ signalling pathway. J Cell Mol Med 2021;25:4608-22. [Crossref] [PubMed]
- Stark Z, Savarirayan R. Osteopetrosis. Orphanet J Rare Dis 2009;4:5. [Crossref] [PubMed]
- Sugatani T, Hruska KA. Impaired micro-RNA pathways diminish osteoclast differentiation and function. J Biol Chem 2009;284:4667-78. [Crossref] [PubMed]
(English Language Editors: L. Huleatt and J. Chapnick)


