
Identification of key genes in cutaneous squamous cell carcinoma: a transcriptome sequencing and bioinformatics profiling study
Introduction
Cutaneous squamous cell carcinoma (cSCC) is one of the most common malignant cancers worldwide and is characterized by aberrant proliferation of keratinocytes (1). cSCC represents 20% of all non-melanoma skin cancers and is a significant cause of mortality due to its ability to metastasize to any organ in the body and its increasing prevalence (2). Actinic keratosis (AK) is one of the most common skin diseases in the elderly population and is generally considered a precancerous lesion of cSCC (3,4). Many studies have found that AK and cSCC exhibit highly similar clinical, histological, and molecular characteristics (5), and it is estimated that 10% of AK will develop into cSCC in approximately 2 years (6). The most important risk factor for cSCC and AK is exposure to ultraviolet B (UVB) radiation (7). UV radiation is considered a “complete carcinogen” because it is not only a mutagen but also a nonspecific damaging agent and has properties of both a tumor initiator and a tumor promoter (8). UV can cause cellular DNA damage and genetic mutations due to its powerful genotoxic effects (9,10). In addition, UV can induce inflammatory responses (11,12), promote the proliferation of epidermal keratinocytes (13), and cause immune tolerance or immunosuppression (14).
Studies have detailed the multiple mechanisms involved in the incidence and development of cSCC, including critical gene mutations (e.g., TP53, NOTCH 1-2, CDKN2A, FOS, RAS, EGFR, MYC, and BRAF) (15-18), key signaling pathway changes (e.g., PI3K/Akt, TGF-β, MAPK, NF-κB, and WNT) (19-22), immunodeficiency (23), microenvironment changes (24), epithelial-mesenchymal transition (EMT) (25), altered expression of adhesion molecules, reorganization of cytoskeletal proteins, and production of extracellular matrix (ECM)-degrading enzymes (26). However, skin cancer is a complex process, and the precise mechanisms involved in cSCC remain to be fully elucidated. Given the systemic metastatic and recurrent nature of cSCC (27), and the increasing incidence of AK and cSCC, more research is needed to identify key molecules involved in the transition of AK to cSCC, and the invasion and metastasis of cSCC. Furthermore, previous studies (28,29) examining the molecular pathogenesis of cSCC has primarily focused on fair-skinned populations in Europe, with few studies in Asian populations. Therefore, in this current study, transcriptome sequencing was conducted on 24 individuals from the plateau region of Yunnan, China, where UV causes a high incidence of AK and cSCC. Transcriptome sequencing and bioinformatics analysis can efficiently evaluate whole processes in one tissue in a global manner. Four types of tissues were examined, including AK, cSCC, normal sun-exposed skin (NES), and normal non-sun-exposed skin (NNS), and the differentially expressed genes (DEGs) were identified and further validated by real-time quantitative reverse transcription PCR (qRT-PCR). The results from this study provide an improved understanding of the occurrence and development of cSCC, and may provide helpful insights into early clinical defense mechanisms and effective treatment strategies for the management of AK and cSCC.
We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/atm-21-3915).
Methods
Tissue samples
Tissue samples of NNS (n=6), NES (n=6), AK (n=6), and cSCC (n=6) were obtained from patients during surgical excision at the Dermatology Department of the First Affiliated Hospital of Kunming Medical University (Yunnan, China). All AK and cSCC samples were derived from the face, while NES and NNS samples were derived from the face and trunk of healthy participants who were undergoing nevus resection surgery. Half of each sample was immediately snap-frozen in liquid nitrogen and stored at −80 °C for RNA extraction and gene expression analysis. The other half was fixed in formalin, embedded in paraffin, and used for histopathological diagnosis by two independent pathologists. Patients were aged 72.22±4.66 years (range, 65–78 years old). There were no significant differences in gender or age distribution among the four groups, and none of these patients had received any form of treatment prior to surgery. This study protocol was approved by the Ethics Committee of the First Affiliated Hospital of Kunming Medical University [Approval Number (2020)-L-29], and written informed consent was obtained from all patients. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
RNA sequencing (RNA-Seq)
Total RNA extraction was performed using TRIzol reagent (TaKaRa Bio Inc., Kusatsu, Japan) following the manufacturer’s protocol, and RNA degradation and contamination were assessed by 1% agarose gel electrophoresis. Sequencing libraries were generated using the NEBNext®UltraTM RNA Library Prep Kit for Illumina® (NEB, USA), and library fragments were purified using the AMPure XP system (Beckman Coulter, Beverly, USA) to select cDNA fragments of the right length for PCR. Products were purified (AMPure XP system) and library quality was assessed on an Agilent Bioanalyzer 2100 system and sequenced on the Illumina HiSeq 4000 platform, and 150 bp paired-end reads were generated.
Bioinformatics analysis
The high-quality clean paired-end reads were aligned to the reference genome using Hisat2 software (http://ccb.jhu.edu/software/hisat2). The total number of mapped reads was determined, and the FPKMs (fragments per kilobase per million mapped reads) were calculated for each annotated gene. The R software package DESeq2 was employed to capture the DEGs and calculate the fold changes for each gene. Genes with|log2Fold change|>2, false discovery rate (FDR) <0.05, and Padj <0.05 were defined as DEGs and utilized for further analysis. In addition, the EMT-related genes (ERGs) and autophagy-related genes (ARGs) were obtained from public databases (http://dbemt.bioinfo-minzhao.org/index.html; http://www.tanpaku.org/autophagy/list/GeneList.html) to identify differentially expressed ERGs (DEERGs) and ARGs (DEARGs).
Function and pathway enrichment analysis
To analyze the identified DEGs at the functional level, significant Gene Ontology (GO, http://www.geneontology.org) biological process terms and Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg) were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov) with the thresholds of Padj <0.05 and FDR <0.05. To identify pathways of intersecting genes, KEGG enrichment analysis was implemented. ClusterProfiler was applied to visualize the results.
Modules from the protein-protein interaction (PPI) network
To evaluate the interactive relationships among DEGs, the DEGs were mapped to the Search Tool for the Retrieval of Interacting Genes (STRING) database (https://string-db.org). Interactive DEGs were selected to construct the PPI network (combined score ≥0.75) and visualized using Cytoscape. The Molecular Complex Detection (MCODE) plugin in Cytoscape was used to screen the modules of the PPI network with MCODE score >3 and number of nodes ≥2.
Real-time quantitative reverse transcription PCR (qRT-PCR)
qRT-PCR was performed on an independent set of biopsies (n=5 in each group). Five significant DEGs were selected for qRT-PCR analysis, including TCN1, HEPHL1, FBN2, SULF1, SULF2, COL10A1, AREG, and ACTA1. Primers were designed and are listed in Table 1. Total RNA was isolated using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and reverse transcribed to cDNA using an mRNA Reverse Transcription Kit (Invitrogen; Thermo Fisher Scientific, Inc.). qRT-PCR was performed using a SYBR-Green kit (Invitrogen; Thermo Fisher Scientific, Inc.). The RNA expression level of target genes was evaluated using 2−∆∆Ct.
Table 1
| Gene | Forward primer 5'-3' | Reverse primer 5'-3' |
|---|---|---|
| FBN2 | TGAGTGCAGCATCATTCCTGG | AGAAACACATGCCTGTTCTCTG |
| HEPHL1 | GAAAAACTACACCTACGTCTGGC | GATGTGCGAATGGTACACCCA |
| SULF1 | TGGCGAGAATGGCTTGGATTA | TAACGGGCCTATGGGGATACA |
| SULF2 | GGCAGGTTTCAGAGGGACC | GAAGGCGTTGATGAAGTGCG |
| TCN1 | ATTGTCAGATGTAAGCTCGGGA | GCCATTGTGTGCTTCCATATTTT |
| GAPDH | GGACCTGACCTGCCGTCTAG | GTAGCCCAGGATGCCCTTGA |
Statistical analysis
Statistical analyses were performed using SPSS Statistics 24.0 (IBM SPSS Statistics for Windows; IBM Corp, NY, USA). The data are presented as the means ± standard deviations. Differences among groups were analyzed using one-way analysis of variance (ANOVA) for normally distributed data and the Kruskal-Wallis test for non-normally distributed data. P<0.05 was considered statistically significant.
Results
Identification of the differentially expressed genes
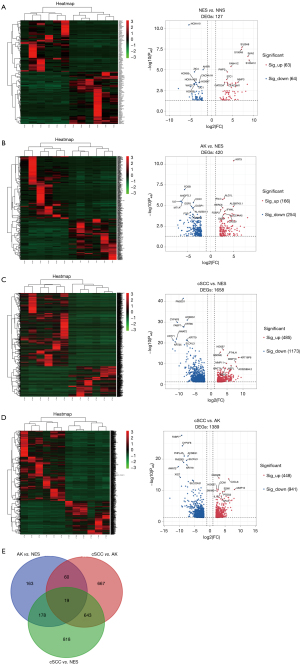
The DEGs in the cSCC, AK, NES, and NNS samples were screened (https://cdn.amegroups.cn/static/public/atm-21-3915-1.xlsx). According to the screening criteria, a total of 127 DEGs (63 upregulated and 64 downregulated) were identified when comparing NES and NNS (Figure 1A), 420 DEGs (166 upregulated and 254 downregulated) were identified when comparing AK and NES samples (Figure 1B), 1,658 DEGs (485 upregulated and 1,173 downregulated) were identified when comparing cSCC and NES (Figure 1C), and 1,389 DEGs (448 upregulated and 941 downregulated) were identified when assessing cSCC and AK samples (Figure 1D). To identify the differentially expressed genes that were co-expressed in cSCC and AK, the DEGs detected during the following comparisons were overlapped: AK vs. NES, cSCC vs. NES, and AK vs. cSCC. As a result, 19 co-expressed DEGs were found (Figure 1E), including KRT32, KRT35, KRT85, CXCL3, CXCL11, ZNF536, CHIT1, F5, ACTA1, MUC16, CADM2, CSF3, CLVS2, SFRP4, LRRTM1, IL6, GFRA3, LINC02577, and EXTL1.
Functional and pathway enrichment analysis of DEGs
The top 10 significant terms from the GO analysis are illustrated in https://cdn.amegroups.cn/static/public/atm-21-3915-2.xlsx. In the biological process (BP) terms, GO analysis results revealed that the upregulated DEGs in the NES vs. NNS analysis were primarily enriched in cell chemotaxis, antimicrobial humoral response, granulocyte chemotaxis and migration, and humoral immune response; while the downregulated DEGs were primarily enriched in the pattern specification process, regionalization, anterior/posterior pattern specification, embryonic skeletal system development and morphogenesis of a branching epithelium. In the AK vs. NES analysis, the upregulated DEGs were enriched in the regulation of lymphocyte activation, humoral immune response, regulation of chronic inflammatory response, and defense response to virus; while downregulated DEGs were enriched in the regulation of the ERK1 and ERK2 cascade and cellular response to metal ion. In the cSCC vs. NES analysis, upregulated DEGs were primarily enriched in the complement activation classical pathway and humoral immune response; and the downregulated DEGs were primarily enriched in skin development, keratinocyte differentiation, and regulation of membrane potential. In the cSCC vs. AK analysis, upregulated DEGs were primarily enriched in cell chemotaxis, skin development, endodermal cell differentiation, and connective tissue development; while downregulated DEGs were primarily enriched in keratinization, fatty acid metabolic process, regulation of lymphocyte activation, and lymphocyte and mononuclear cell proliferation.
For GO cell component (CC), upregulated DEGs in the NES vs. NNS analysis were primarily enriched in the cytoplasmic vesicle lumen, cell membrane, and specific granule; while downregulated DEGs were primarily enriched in the Wnt signalosome. In the AK vs. NES analysis, upregulated DEGs were primarily enriched on the external side of the plasma membrane and immunoglobulin complex; and downregulated DEGs were primarily enriched in the transcription factor AP-1 complex and collagen-containing extracellular matrix. For the cSCC vs. NES analysis, upregulated DEGs were primarily enriched in the extracellular matrix component, complex of collagen trimers, and basement membrane; and downregulated DEGs were primarily enriched in the intermediate filament cytoskeleton and transmembrane transporter complex. In the cSCC vs. AK analysis, upregulated DEGs were primarily enriched in the collagen-containing extracellular matrix, extracellular matrix component, endoplasmic reticulum lumen, basement membrane, and collagen trimer; while downregulated DEGs were primarily enriched in the intermediate filament, intermediate filament cytoskeleton, and keratin filament.
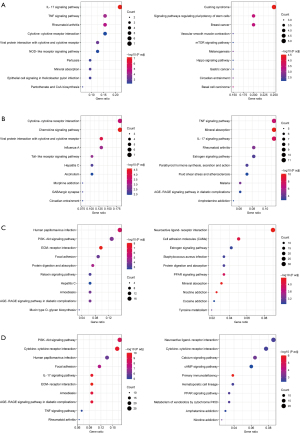
KEGG pathway enrichment analysis was used to identify the canonical signaling pathways associated with the significant DEGs. In the NES vs. NNS comparison, 42 significant KEGG pathways were annotated (Figure 2A). The upregulated DEGs were primarily involved in the interleukin (IL)-17 signaling pathway, tumor necrosis factor (TNF) signaling pathway, and cytokine-cytokine receptor interaction; while downregulated DEGs were primarily involved in signaling pathways regulating the pluripotency of stem cells, mTOR signaling pathway, Hippo signaling pathway, and Wnt signaling pathway. In the AK vs. NES comparison, 69 significant KEGG pathways were annotated (Figure 2B). The upregulated DEGs were primarily involved in the chemokine signaling pathway, viral protein interaction with cytokine and cytokine receptor, cytokine-cytokine receptor interaction, and Toll-like receptor signaling pathway; while downregulated DEGs were primarily involved in the IL-17 signaling pathway, TNF signaling pathway, and NOD-like receptor signaling pathway. In the cSCC vs. NES comparison, 60 significant KEGG pathways were annotated (Figure 2C). The upregulated DEGs were primarily involved in ECM-receptor interaction, human papillomavirus infection, PI3K-Akt signaling pathway, focal adhesion, mucin type O-glycan biosynthesis, regulating pluripotency of stem cells, and Wnt signaling pathway; while the downregulated DEGs were primarily involved in cell adhesion molecules (CAMs), PPAR signaling pathway, tyrosine metabolism, and cAMP signaling pathway. In the cSCC vs. AK analysis, 78 significant KEGG pathways were annotated (Figure 2D). The upregulated DEGs were mainly involved in ECM-receptor interaction, IL-17 signaling pathway, cytokine-cytokine receptor interaction, PI3K-Akt signaling pathway, focal adhesion, human papillomavirus infection, and TNF signaling pathway; and the downregulated DEGs were mainly associated with immunodeficiency and calcium signaling pathways.
Protein-protein (PPI) network construction
Protein interactions among the DEGs were predicted using the STRING online database and Cytoscape software. A total of 37 nodes and 47 edges were involved in the PPI network of the DEGs identified in the NES vs. NNS analysis, 134 nodes and 333 edges were involved in the DEGs identified in the AK vs. NES analysis, 600 nodes and 2,618 edges were involved in the DEGs identified in the cSCC vs. NES comparison, and 462 nodes and 1647 edges were involved in the DEGs identified in the cSCC vs. AK comparison (https://cdn.amegroups.cn/static/public/atm-21-3915-3.xlsx). The results revealed that genes in the keratin family and the chemotactic cytokine family, including KRT6A, KRT6B, KRT6C, KRT16, KRT17, KRT28, KRT31, KRT32, KRT35, KRT79, KRT83, KRT85 and CXCL3, CXCL5, CXCL8, and CXCL11, were the most significantly altered genes in NES, AK, and cSCC tissues compared to NNS tissues.
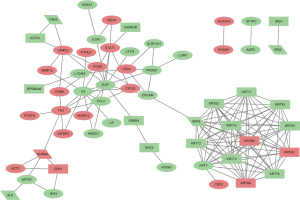
Accumulating evidence has shown that EMT and autophagy play critical roles in cSCC (30,31). To understand the EMT and autophagy-related genes involved in the occurrence of cSCC, a total of 1,011 ERGs and 742 ARGs were screened from the RNA-Seq data and public databases and PPI maps were constructed for the significant DEERGs and DEARGs in cSCC compared to NES. The final PPI network complex contained 59 nodes and 141 edges (Figure 3).
Validation of significant differentially expressed genes using qRT-PCR
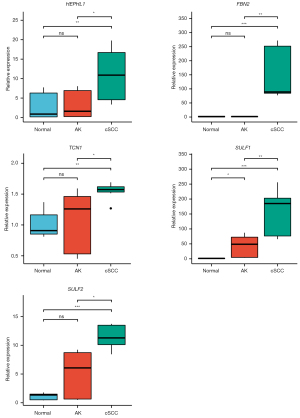
To validate the accuracy of the RNA sequencing results, qRT-PCR was performed on 5 significant DEGs in cSCC tissues, AK tissues, and NES tissues from 5 different patients. As shown in Figure 4, the expression of HEPHL1, FBN2, SULF1, SULF2, and TCN1 was significantly upregulated in cSCC samples compared to NES and AK tissues, which was consistent with the RNA-Seq results.
Discussion
Although previous profiling experiments in cSCC have identified a large number of DEGs, the field has been hampered by a lack of consensus among these studies (3,32,33). Therefore, more in-depth studies are needed. Even as new sequencing technologies continue to evolve, transcriptome sequencing techniques still play a major role in identifying cancer-specific genes. In this study, next-generation sequencing technology was used to analyze the DEGs among NNS, NES, AK, and cSCC tissues from patients in Yunnan, China. A group of 5 DEGs that were highly associated with AK and cSCC tissues was identified and validated.
The gene expression patterns or the transcriptomes of cancer patients are greatly impacted by multiple factors, including infected microbiomes (34,35) and the severity of the disease (36), and thus, there can be considerable interpatient variation. Indeed, this study demonstrated that the expression levels of DEGs were significantly different between cSCC patients. Furthermore, the statistical results of all the DEGs in our study showed that each patient has a unique pattern of gene expression. The expression patterns of many DEGs in the transcriptome were not stable, which may be due to the heterogeneity of the tumor, the development of the disease, or the genetic background of the individual (37,38). This is a hindrance for researchers attempting to identify cancer driver genes for cSCC. Therefore, individual differences must be considered when conducting subsequent studies. Single-cell transcriptome sequencing (scRNA-seq) is a new technology for high-throughput sequencing analysis at the level of the single cell and may be a promising tool for exploring the heterogeneity and development of skin cancer (39).
By analyzing the overall changes in gene expression, the degree of DEG expression was most evident in AK and cSCC tissues, whereas normal skin appeared to have minimal alterations in gene expression. Based on human samples obtained from non-sun-exposed and sun-exposed skin, a small number of significant DEGs was identified. Among the top 20 significant DEGs, the S100 gene family (S100A8, S100A9, S100A12) and the SAA gene family (SAA1, SAA2) were significantly upregulated, while the HOX gene family (HOXA10, HOXB5, HOXB7, HOXA-AS3) was significantly downregulated. The S100 gene family plays an important role in the response to environmental triggers and cellular damage by regulating cellular differentiation, proliferation, apoptosis, inflammation, calcium homeostasis, cytoskeleton, and microbial resistance (40). S100 family proteins are mainly expressed in epidermal and skin tumors (41), among which S100A8 and S100A9 are considered to be key pro-inflammatory factors (42). S100A8 and S100A9 are secreted by psoriatic keratinocytes, and then send signals to adjacent keratinocytes through positive feedback to produce pro-inflammatory and pro-angiogenic cytokines, which exacerbate psoriasis skin lesions without affecting proliferation of keratinocytes. S100A8, S100A9, and S100A12 are positively correlated with cSCC tumorigenesis and highly expressed in cSCC. They can play an important role in the invasiveness of cSCC by enhancing in vitro proliferation and migration (41,43), which is consistent with our findings. The protein expression of the SAA gene is induced during acute and chronic inflammation or in response to tissue injury (44). Currently, SAA has been used as a predictor of cancer risk in various human malignancies. SAA is produced by the liver and enters the systemic circulation in response to the stimulation of inflammatory cytokines (such as IL-6) and participates in cholesterol transport, extracellular matrix degradation and inflammatory cell recruitment. The increase in SAA may be caused by too many inflammatory cytokines, especially IL-6. At present, not only has SAA highly expressed in a variety of cancer tissues, but also SAA gene families in AK and cSCC tissues are also significantly up-regulated in this experiment, indicating that elevated SAA levels may also be the main product of cancer (45,46). HOX genes encode a highly evolutionarily conserved transcription factor family known to be key regulators of embryonic development and various cellular processes, including cell growth, differentiation, apoptosis, motility, and angiogenesis (47). It can act as a tumor promoter or suppressor. Other DEGs, such as MMP3, CXCL5, CXCL8, CCL20, and CXCL1, were primarily enriched in inflammatory and immune-related pathways in AK and cSCC (48,49). Normal inflammation is self-limiting because the production of anti-inflammatory cytokines closely follows pro-inflammatory cytokines, while the subversion of cell death and/or repair processes occurs in chronically inflamed tissues, leading to DNA replication and loss of normal cell proliferation and growth control. Previous data demonstrated that the inflammatory response is a critical player in the tumor process, promoting the proliferation, survival, and migration of tumor cells (50). Therefore, the difference in the expression of DEG in AK and cSCC tissues suggests that inflammation may mediate the occurrence of the disease and occupy an important position.
The finding of similar DEGs between AK and cSCC tissues confirmed that AK is a precursor lesion of cSCC and indicated that they are genetically closely related. Nearly half of the DEGs identified in the AK vs. NES analysis overlapped with the DEGs identified in the cSCC vs. NES analysis. ECM-receptor interaction pathways were the most upregulated gene-enriched signaling pathways identified in the cSCC vs. NES analysis and the cSCC vs. AK analysis. The extracellular matrix (ECM) is reported to constitute the scaffold of tumor microenvironment and regulate cancer behavior (51). ECM-receptor interaction pathways were the most upregulated gene-enriched signaling pathways that play an important role in the process of tumor shedding, adhesion, degradation, movement and hyperplasia. The role of ECM in other cancers has been proved. ECM is upregulated in breast cancer tissue (52) and participates in the process of tumor invasion and metastasis in gastric cancer (53). Dysregulation of ECM remodeling caused by cancer cells promotes irreversible protein hydrolysis and cross-linking, which in turn affects cell signaling, microenvironmental cues, angiogenesis, and tissue biomechanics (54). Matrix metalloproteinases (MMPs) are an important example. MMPs are zinc-containing endopeptidases with an extensive range of substrate specificities. These enzymes are able to degrade various components of ECM proteins and are involved in angiogenesis, which promotes cancer cell growth and migration (55). In our results, increased expression of MMP13, MMP10, and MMP1 was observed in cSCC samples, which was consistent with previous reports (56,57). This indicated that MMP might affect the ECM-receptor interaction pathway by degrading various ECM proteins to promote the occurrence and development of tumors. Microorganisms exist on the surface of skin and participate in various physiological processes. All microorganisms living in the human body are an indispensable part of maintaining health and immune system (58). Recent studies demonstrate correlations between a particular composition of microbiome and a broad spectrum of diseases, including lung cancer, obesity, and even breast cancer (59,60). Microbiome-related pathways were also enriched in both AK and cSCC, especially the human papilloma virus (HPV), which has been shown to be positively correlated with keratinocyte cell proliferation, inflammation, and the immune response (34,61), suggesting that HPV is closely related to the occurrence of AK and cSCC and the transformation of AK to cSCC (62-64). In addition, a group of EMT- and autophagy-related genes involved in cSCC was identified, and most of the DEGs related to autophagy were downregulated. These results suggested that inhibition of autophagy and activation of EMT plays an important role in the occurrence and development of cSCC.
Moreover, 5 highly expressed DEGs were confirmed by qRT-PCR in the cSCC vs. NES analysis and the cSCC vs. AK analysis. Both hephaestin-like 1 (HEPHL1), which is thought to function as a ferroxidase and facilitates the transfer of iron to transferrin, and HMOX1, were highly expressed in cSCC. Only a few studies have focused on the role of HEPHL1 in tumors. HEPHL1 mutation has been detected in POEMS syndrome (65) and may also be related to the malignant transformation of hepatocyte L02 cells induced by microcystin-LR (66). Therefore, what role HEPHL1 plays in cSCC needs to be further studied. Similar to HEPHL1, FBN2, TCN1, SULF1, and SULF2 were also highly expressed in cSCC. Fibrillin 2 (FBN2) encodes a component of connective tissue microfibrils and forms the structural core of microfibrils. Previous studies have shown that methylation or silencing of FBN2 in tumor cells may play an important role in carcinogenesis, invasion, and metastasis (67). However, recent research suggested the opposite, wherein FBN2 stimulates tumor angiogenesis by influencing TGF-β sequestration by microfibrils, leading to a locally higher active TGF-β concentration in the tumor microenvironment which is involved in tumor progression, metastasis, EMT, and tumor angiogenesis (68). In lung cancer, cholangiocarcinoma, and metastatic colorectal cancer, FBN2 has been found to be associated with EMT, cell migration, and metastasis (69-71). In combination with our KEGG pathway enrichment analysis, we found that the ECM-receptor interaction pathway was significantly up-regulated and the expression of MMP protein increased. We speculate that FBN2 may be their inducing factor, and affect the tumor microenvironment by acting on them. Transcobalamin I (TCN1) encodes a member of the vitamin B12-binding protein family, is expressed in various tissues and secretions, and regulates cobalamin homeostasis. Studies have shown that TCN1 is associated with poor outcomes in malignancies, such as pancreatic and colon cancer (72,73). Sulfatase 1 (SULF1) and sulfatase 2 (SULF2) encode extracellular heparan sulfate endosulfatase, which can edit the sulfation status of heparan sulfate proteoglycans (HSPGs) on the outside of cells and regulate a number of critical signaling pathways, such as Wnt/β-catenin signaling (74). Recent studies have shown that SULF1 and SULF2 are involved in the occurrence and development of many tumors, including breast cancer, lung cancer, and prostate cancer, indicating their potential for cancer diagnosis and therapy (75-77). SULF1 is highly expressed in colon cancer, and an in vitro study confirmed that M2-like macrophages in colon cancer increased the amount of SULF1 produced by fibroblasts and were associated with poor prognosis (78). Graham et al. (79) found that the activation of SULF1/SULF2 not only promoted fetal growth regulation and injury-induced liver regeneration but also dysregulated tumor growth. Interestingly, SULF1 and SULF2 were significantly elevated in cSCC compared to AK and normal skin in our study, indicating that the SULF family plays an important role in cSCC and may mediate tumor growth.
Interestingly, the expression of ACTA1 in NES, AK, and cSCC tissues exhibited a downward trend as the disease progressed. ACTA1 is downregulated in head and neck squamous cell carcinoma and may serve as a potential diagnostic and prognostic marker of tumors (80). A similar result was found in colorectal cancer and pancreatic adenocarcinoma (81). A large amount of research shows that keratin is associated with a variety of diseases (82,83). In the current study, the keratin gene family, including KRT 85, KRT 32, and KRT 35, displayed the same downward trend as ACTA1. Actin and keratin are known to be components of the cytoskeleton (84) (a schematic diagram of the cytoskeleton is shown in Figure 5). The various components of the cytoskeleton are well orchestrated in normal cells, and mutations and abnormal expression of cytoskeletal and cytoskeletal-associated proteins play an important role in cancer cell migration, invasion, and metastasis. The actin family of proteins are highly conserved proteins that play a role in cell motility, structure, and integrity. Studies have shown that actin cytoskeletal rearrangement is associated with invasion and metastatic phenotypes of various cancer cells (85-87). Furthermore, upregulation of the keratin family KRT80 can promote cytoskeletal rearrangements at the leading edge and increase focal adhesion and cellular stiffening, collectively promoting cancer cell invasion that is associated with poor survival (88). High expression of KRT80 in gastric cancer and colorectal cancer is also involved in the malignant proliferation of tumors (89,90). A study of basal cell carcinoma (BCC) showed a close correlation between differentiation‐specific keratins and BCC (91). Our study found that keratinocyte differentiation and hyperproliferation-related genes, and stress markers (e.g., KRT6A, KRT6B, KRT6C, KRT14, and KRT16) were upregulated in cSCC. Some members of the keratin family (KRT32, KRT35, KRT85) were significantly and consistently downregulated in AK and cSCC compared to normal skin. This could suggest that certain types of keratin may play different roles in the progression of cancer, especially in the migration, invasion and metastasis of cancer cells.
CXCL11 is also highly expressed in cSCC, and studies have shown that it can bind to CXCR3 and CXCR7, which is associated with angiogenesis, invasiveness, and reduction of apoptosis in tumor cells (92,93). GFRA3 is a glycosylphosphatidylinositol (GPI)-linked cell surface receptor. Eftang et al. (94) found that high methylation levels of GFRA3 conferred a very unfavorable prognosis in gastric cancer. However, expression of GFRA3 was associated with the growth of cancer cells, tumor lymph node metastases, and higher clinical stages of breast cancer and non-small cell lung cancer (95,96). These studies suggested that GFRA3 may play different roles in various cancer types. In our study, GFRA3 was downregulated and may be related to antitumor properties, but the exact mechanisms remain to be further studied.
Conclusions
The molecular characterization of AK and cSCC is an important step towards identifying and understanding the dysregulated pathways and key drivers of cancer development and progression. This study identified some important genes that may play crucial roles in the occurrence and development of cSCC, and perhaps AK. To the best of our knowledge, this is the first study to examine the function of these genes in cSCC. Additional studies should be performed to further explore the pathogenic mechanisms of these genes.
Acknowledgments
The authors acknowledge the editors and reviewers for their positive and constructive comments and suggestions related to this study.
Funding: This work was supported by the Yunnan Science and Technology Leading Talents Project (grant number 2017HA010), the Yunnan Province Clinical Research Center for Skin Immune Diseases (grant number 2019ZF012), and the Yunnan Province Clinical Center for Skin Immune Diseases (grant number ZX2019-03-02).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-3915
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-3915
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-3915). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). This study protocol was approved by the Ethics Committee of the First Affiliated Hospital of Kunming Medical University [Approval Number (2020)-L-29], and written informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Parekh V, Seykora JT. Cutaneous Squamous Cell Carcinoma. Clin Lab Med 2017;37:503-25. [Crossref] [PubMed]
- Burton KA, Ashack KA, Khachemoune A. Cutaneous Squamous Cell Carcinoma: A Review of High-Risk and Metastatic Disease. Am J Clin Dermatol 2016;17:491-508. [Crossref] [PubMed]
- Hameetman L, Commandeur S, Bavinck JN, et al. Molecular profiling of cutaneous squamous cell carcinomas and actinic keratoses from organ transplant recipients. BMC Cancer 2013;13:58. [Crossref] [PubMed]
- Mittelbronn MA, Mullins DL, Ramos-Caro FA, et al. Frequency of pre-existing actinic keratosis in cutaneous squamous cell carcinoma. Int J Dermatol 1998;37:677-81. [Crossref] [PubMed]
- Filosa A, Filosa G. Actinic keratosis and squamous cell carcinoma: clinical and pathological features. G Ital Dermatol Venereol 2015;150:379-84. [PubMed]
- Fuchs A, Marmur E. The kinetics of skin cancer: progression of actinic keratosis to squamous cell carcinoma. Dermatol Surg 2007;33:1099-101. [Crossref] [PubMed]
- Leiter U, Keim U, Garbe C. Epidemiology of Skin Cancer: Update 2019. Adv Exp Med Biol 2020;1268:123-39. [Crossref] [PubMed]
- D'Orazio J, Jarrett S, Amaro-Ortiz A, et al. UV radiation and the skin. Int J Mol Sci 2013;14:12222-48. [Crossref] [PubMed]
- Pfeifer GP, You YH, Besaratinia A. Mutations induced by ultraviolet light. Mutat Res 2005;571:19-31. [Crossref] [PubMed]
- Cleaver JE, Crowley E. UV damage, DNA repair and skin carcinogenesis. Front Biosci 2002;7:d1024-43. [PubMed]
- Matsumura Y, Ananthaswamy HN. Toxic effects of ultraviolet radiation on the skin. Toxicol Appl Pharmacol 2004;195:298-308. [Crossref] [PubMed]
- Clydesdale GJ, Dandie GW, Muller HK. Ultraviolet light induced injury: immunological and inflammatory effects. Immunol Cell Biol 2001;79:547-68. [Crossref] [PubMed]
- Bayerl C, Taake S, Moll I, et al. Characterization of sunburn cells after exposure to ultraviolet light. Photodermatol Photoimmunol Photomed 1995;11:149-54. [Crossref] [PubMed]
- Kripke ML. Reflections on the field of photoimmunology. J Invest Dermatol 2013;133:27-30. [Crossref] [PubMed]
- Zhao L, Li W, Marshall C, et al. Srcasm inhibits Fyn-induced cutaneous carcinogenesis with modulation of Notch1 and p53. Cancer Res 2009;69:9439-47. [Crossref] [PubMed]
- Inman GJ, Wang J, Nagano A, et al. The genomic landscape of cutaneous SCC reveals drivers and a novel azathioprine associated mutational signature. Nat Commun 2018;9:3667. [Crossref] [PubMed]
- Missero C. The genetic evolution of skin squamous cell carcinoma: tumor suppressor identity matters. Exp Dermatol 2016;25:863-4. [Crossref] [PubMed]
- Martincorena I, Roshan A, Gerstung M, et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 2015;348:880-6. [Crossref] [PubMed]
- Xu D, Yuan R, Gu H, et al. The effect of ultraviolet radiation on the transforming growth factor beta 1/Smads pathway and p53 in actinic keratosis and normal skin. Arch Dermatol Res 2013;305:777-86. [Crossref] [PubMed]
- Zhang J, Jiang H, Xu D, et al. DNA-PKcs Mediates An Epithelial-Mesenchymal Transition Process Promoting Cutaneous Squamous Cell Carcinoma Invasion And Metastasis By Targeting The TGF-β1/Smad Signaling Pathway. Onco Targets Ther 2019;12:9395-405. [Crossref] [PubMed]
- Han S, Li X, Liang X, et al. HOXA9 Transcriptionally Promotes Apoptosis and Represses Autophagy by Targeting NF-κB in Cutaneous Squamous Cell Carcinoma. Cells 2019;8:1360. [Crossref]
- Chu Z, Zhang X, Li Q, et al. CDC20 contributes to the development of human cutaneous squamous cell carcinoma through the Wnt/β‑catenin signaling pathway. Int J Oncol 2019;54:1534-44. [Crossref] [PubMed]
- Bottomley MJ, Thomson J, Harwood C, et al. The Role of the Immune System in Cutaneous Squamous Cell Carcinoma. Int J Mol Sci 2019;20:2009. [Crossref] [PubMed]
- Nissinen L, Farshchian M, Riihilä P, et al. New perspectives on role of tumor microenvironment in progression of cutaneous squamous cell carcinoma. Cell Tissue Res 2016;365:691-702. [Crossref] [PubMed]
- Hodorogea A, Calinescu A, Antohe M, et al. Epithelial-Mesenchymal Transition in Skin Cancers: A Review. Anal Cell Pathol (Amst) 2019;2019:3851576 [Crossref] [PubMed]
- Wang X, Enomoto A, Weng L, et al. Girdin/GIV regulates collective cancer cell migration by controlling cell adhesion and cytoskeletal organization. Cancer Sci 2018;109:3643-56. [Crossref] [PubMed]
- Que SKT, Zwald FO, Schmults CD. Cutaneous squamous cell carcinoma: Incidence, risk factors, diagnosis, and staging. J Am Acad Dermatol 2018;78:237-47. [Crossref] [PubMed]
- Ioannidis NM, Wang W, Furlotte NA, et al. Gene expression imputation identifies candidate genes and susceptibility loci associated with cutaneous squamous cell carcinoma. Nat Commun 2018;9:4264. [Crossref] [PubMed]
- Sordillo JE, Kraft P, Wu AC, et al. Quantifying the Polygenic Contribution to Cutaneous Squamous Cell Carcinoma Risk. J Invest Dermatol 2018;138:1507-10. [Crossref] [PubMed]
- Fernandez-Figueras MT, Puig L. The Role of Epithelial-to-Mesenchymal Transition in Cutaneous Squamous Cell Carcinoma : Epithelial-to-Mesenchymal Transition in Cutaneous SCC. Curr Treat Options Oncol 2020;21:47. [Crossref] [PubMed]
- Sample A, He YY. Autophagy in UV Damage Response. Photochem Photobiol 2017;93:943-55. [Crossref] [PubMed]
- Yuan SP, Li CX, Qin S, et al. High expression of disabled homolog 2-interacting protein contributes to tumor development and proliferation in cutaneous squamous cell carcinoma. Ann Transl Med 2020;8:1131. [Crossref] [PubMed]
- Van Haren R, Feldman D, Sinha AA. Systematic comparison of nonmelanoma skin cancer microarray datasets reveals lack of consensus genes. Br J Dermatol 2009;161:1278-87. [Crossref] [PubMed]
- Kullander J, Forslund O, Dillner J. Staphylococcus aureus and squamous cell carcinoma of the skin. Cancer Epidemiol Biomarkers Prev 2009;18:472-8. [Crossref] [PubMed]
- Mallika L, Augustine D, Rao RS, et al. Does microbiome shift play a role in carcinogenesis? A systematic review. Transl Cancer Res 2020;9:3153-66. [Crossref]
- Spiegel D, Giese-Davis J. Depression and cancer: mechanisms and disease progression. Biol Psychiatry 2003;54:269-82. [Crossref] [PubMed]
- Gowthami J, Gururaj N, Mahalakshmi V, et al. Genetic predisposition and prediction protocol for epithelial neoplasms in disease-free individuals: A systematic review. J Oral Maxillofac Pathol 2020;24:293-307. [Crossref] [PubMed]
- Ponder BA. Cancer genetics. Nature 2001;411:336-41. [Crossref] [PubMed]
- Zou D, Qi J, Wu W, et al. Applications of Single-Cell Sequencing in Dermatology. Med Sci Monit 2021;27:e931862 [Crossref] [PubMed]
- Sreejit G, Flynn MC, Patil M, et al. S100 family proteins in inflammation and beyond. Adv Clin Chem 2020;98:173-231. [Crossref] [PubMed]
- Choi DK, Li ZJ, Chang IK, et al. Clinicopathological roles of S100A8 and S100A9 in cutaneous squamous cell carcinoma in vivo and in vitro. Arch Dermatol Res 2014;306:489-96. [Crossref] [PubMed]
- Zeng ML, Zhu XJ, Liu J, et al. An Integrated Bioinformatic Analysis of the S100 Gene Family for the Prognosis of Colorectal Cancer. Biomed Res Int 2020;2020:4746929 [Crossref] [PubMed]
- Wei W, Chen Y, Xu J, et al. Identification of Biomarker for Cutaneous Squamous Cell Carcinoma Using Microarray Data Analysis. J Cancer 2018;9:400-6. [Crossref] [PubMed]
- Zhang Y, Zhang J, Sheng H, et al. Acute phase reactant serum amyloid A in inflammation and other diseases. Adv Clin Chem 2019;90:25-80. [Crossref] [PubMed]
- Wang JY, Zheng YZ, Yang J, et al. Elevated levels of serum amyloid A indicate poor prognosis in patients with esophageal squamous cell carcinoma. BMC Cancer 2012;12:365. [Crossref] [PubMed]
- Conti P, Bartle L, Barbacane RC, et al. Synergistic activation of serum amyloid A (SAA) by IL-6 and IL-1 in combination on human Hep 3B hepatoma cell line. Role of PGE2 and IL-1 receptor antagonist. Immunol Invest 1995;24:523-35. [Crossref] [PubMed]
- Contarelli S, Fedele V, Melisi D. HOX Genes Family and Cancer: A Novel Role for Homeobox B9 in the Resistance to Anti-Angiogenic Therapies. Cancers (Basel) 2020;12:3299. [Crossref] [PubMed]
- Liang SB, Ohtsuki Y, Furihata M, et al. Sun-exposure- and aging-dependent p53 protein accumulation results in growth advantage for tumour cells in carcinogenesis of nonmelanocytic skin cancer. Virchows Arch 1999;434:193-9. [Crossref] [PubMed]
- Pacholczyk M, Czernicki J, Ferenc T. The effect of solar ultraviolet radiation (UVR) on induction of skin cancers. Med Pr 2016;67:255-66. [Crossref] [PubMed]
- Coussens LM, Werb Z. Inflammation and cancer. Nature 2002;420:860-7. [Crossref] [PubMed]
- Wu Y, Xu Y. Integrated bioinformatics analysis of expression and gene regulation network of COL12A1 in colorectal cancer. Cancer Med 2020;9:4743-55. [Crossref] [PubMed]
- Bao Y, Wang L, Shi L, et al. Transcriptome profiling revealed multiple genes and ECM-receptor interaction pathways that may be associated with breast cancer. Cell Mol Biol Lett 2019;24:38. [Crossref] [PubMed]
- Yan P, He Y, Xie K, et al. In silico analyses for potential key genes associated with gastric cancer. PeerJ 2018;6:e6092 [Crossref] [PubMed]
- Mohan V, Das A, Sagi I. Emerging roles of ECM remodeling processes in cancer. Semin Cancer Biol 2020;62:192-200. [Crossref] [PubMed]
- Pittayapruek P, Meephansan J, Prapapan O, et al. Role of Matrix Metalloproteinases in Photoaging and Photocarcinogenesis. Int J Mol Sci 2016;17:868. [Crossref] [PubMed]
- Piipponen M, Nissinen L, Riihilä P, et al. p53-Regulated Long Noncoding RNA PRECSIT Promotes Progression of Cutaneous Squamous Cell Carcinoma via STAT3 Signaling. Am J Pathol 2020;190:503-17. [Crossref] [PubMed]
- Farshchian M, Nissinen L, Siljamäki E, et al. Tumor cell-specific AIM2 regulates growth and invasion of cutaneous squamous cell carcinoma. Oncotarget 2017;8:45825-36. [Crossref] [PubMed]
- Lloyd-Price J, Mahurkar A, Rahnavard G, et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 2017;550:61-6. [Crossref] [PubMed]
- Katayama Y, Yamada T, Shimamoto T, et al. The role of the gut microbiome on the efficacy of immune checkpoint inhibitors in Japanese responder patients with advanced non-small cell lung cancer. Transl Lung Cancer Res 2019;8:847-53. [Crossref] [PubMed]
- Reed JP, Devkota S, Figlin RA. Gut microbiome, antibiotic use, and immunotherapy responsiveness in cancer. Ann Transl Med 2019;7:S309. [Crossref] [PubMed]
- Madhusudhan N, Pausan MR, Halwachs B, et al. Molecular Profiling of Keratinocyte Skin Tumors Links Staphylococcus aureus Overabundance and Increased Human β-Defensin-2 Expression to Growth Promotion of Squamous Cell Carcinoma. Cancers (Basel) 2020;12:541. [Crossref] [PubMed]
- Patra V, Gallais Sérézal I, Wolf P. Potential of Skin Microbiome, Pro- and/or Pre-Biotics to Affect Local Cutaneous Responses to UV Exposure. Nutrients 2020;12:1795. [Crossref] [PubMed]
- Paradisi A, Waterboer T, Ricci F, et al. Concomitant seropositivity for HPV 16 and cutaneous HPV types increases the risk of recurrent squamous cell carcinoma of the skin. Eur J Dermatol 2020;30:493-8. [Crossref] [PubMed]
- Chahoud J, Semaan A, Chen Y, et al. Association Between β-Genus Human Papillomavirus and Cutaneous Squamous Cell Carcinoma in Immunocompetent Individuals-A Meta-analysis. JAMA Dermatol 2016;152:1354-64. [Crossref] [PubMed]
- Nagao Y, Mimura N, Takeda J, et al. Genetic and transcriptional landscape of plasma cells in POEMS syndrome. Leukemia 2019;33:1723-35. [Crossref] [PubMed]
- Chen HQ, Zhao J, Li Y, et al. Gene expression network regulated by DNA methylation and microRNA during microcystin-leucine arginine induced malignant transformation in human hepatocyte L02 cells. Toxicol Lett 2018;289:42-53. [Crossref] [PubMed]
- Chen H, Suzuki M, Nakamura Y, et al. Aberrant methylation of FBN2 in human non-small cell lung cancer. Lung Cancer 2005;50:43-9. [Crossref] [PubMed]
- van Loon K, Yemelyanenko-Lyalenko J, Margadant C, et al. Role of fibrillin-2 in the control of TGF-β activation in tumor angiogenesis and connective tissue disorders. Biochim Biophys Acta Rev Cancer 2020;1873:188354 [Crossref] [PubMed]
- Liu ZH, Lian BF, Dong QZ, et al. Whole-exome mutational and transcriptional landscapes of combined hepatocellular cholangiocarcinoma and intrahepatic cholangiocarcinoma reveal molecular diversity. Biochim Biophys Acta Mol Basis Dis 2018;1864:2360-8. [Crossref] [PubMed]
- Kamal Y, Schmit SL, Hoehn HJ, et al. Transcriptomic Differences between Primary Colorectal Adenocarcinomas and Distant Metastases Reveal Metastatic Colorectal Cancer Subtypes. Cancer Res 2019;79:4227-41. [Crossref] [PubMed]
- Hong Q, Li R, Zhang Y, et al. Fibrillin 2 gene knockdown inhibits invasion and migration of lung cancer cells. Cell Mol Biol (Noisy-le-grand) 2020;66:190-6. [Crossref] [PubMed]
- Zhu X, Yi K, Hou D, et al. Clinicopathological Analysis and Prognostic Assessment of Transcobalamin I (TCN1) in Patients with Colorectal Tumors. Med Sci Monit 2020;26:e923828 [Crossref] [PubMed]
- Wu Y, Wei J, Ming Y, et al. Orchestrating a biomarker panel with lncRNAs and mRNAs for predicting survival in pancreatic ductal adenocarcinoma. J Cell Biochem 2018;119:7696-706. [Crossref] [PubMed]
- Yang JD, Sun Z, Hu C, et al. Sulfatase 1 and sulfatase 2 in hepatocellular carcinoma: associated signaling pathways, tumor phenotypes, and survival. Genes Chromosomes Cancer 2011;50:122-35. [Crossref] [PubMed]
- Zhu C, He L, Zhou X, et al. Sulfatase 2 promotes breast cancer progression through regulating some tumor-related factors. Oncol Rep 2016;35:1318-28. [Crossref] [PubMed]
- Vicente CM, Lima MA, Nader HB, et al. SULF2 overexpression positively regulates tumorigenicity of human prostate cancer cells. J Exp Clin Cancer Res 2015;34:25. [Crossref] [PubMed]
- Lui NS, Yang YW, van Zante A, et al. SULF2 Expression Is a Potential Diagnostic and Prognostic Marker in Lung Cancer. PLoS One 2016;11:e0148911 [Crossref] [PubMed]
- Gong W, Li T. Bioinformatical analysis of correlation between sulfatase-1 (SULF1) and prognosis of colon cancer and underlying mechanisms. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2019;35:1008-13. [PubMed]
- Graham K, Murphy JI, Dhoot GK. SULF1/SULF2 reactivation during liver damage and tumour growth. Histochem Cell Biol 2016;146:85-97. [Crossref] [PubMed]
- Yang K, Zhang S, Zhang D, et al. Identification of SERPINE1, PLAU and ACTA1 as biomarkers of head and neck squamous cell carcinoma based on integrated bioinformatics analysis. Int J Clin Oncol 2019;24:1030-41. [Crossref] [PubMed]
- Liu J, Li H, Sun L, et al. Aberrantly methylated-differentially expressed genes and pathways in colorectal cancer. Cancer Cell Int 2017;17:75. [Crossref] [PubMed]
- Omary MB, Coulombe PA, McLean WH. Intermediate filament proteins and their associated diseases. N Engl J Med 2004;351:2087-100. [Crossref] [PubMed]
- Karantza V. Keratins in health and cancer: more than mere epithelial cell markers. Oncogene 2011;30:127-38. [Crossref] [PubMed]
- Fife CM, McCarroll JA, Kavallaris M. Movers and shakers: cell cytoskeleton in cancer metastasis. Br J Pharmacol 2014;171:5507-23. [Crossref] [PubMed]
- Simiczyjew A, Mazur AJ, Dratkiewicz E, et al. Involvement of β- and γ-actin isoforms in actin cytoskeleton organization and migration abilities of bleb-forming human colon cancer cells. PLoS One 2017;12:e0173709 [Crossref] [PubMed]
- Richter C, Mayhew D, Rennhack JP, et al. Genomic Amplification and Functional Dependency of the Gamma Actin Gene ACTG1 in Uterine Cancer. Int J Mol Sci 2020;21:8690. [Crossref] [PubMed]
- Bäder S, Glaubke E, Grüb S, et al. Effect of the actin- and calcium-regulating activities of ITPKB on the metastatic potential of lung cancer cells. Biochem J 2018;475:2057-71. [Crossref] [PubMed]
- Perone Y, Farrugia AJ, Rodríguez-Meira A, et al. SREBP1 drives Keratin-80-dependent cytoskeletal changes and invasive behavior in endocrine-resistant ERα breast cancer. Nat Commun 2019;10:2115. [Crossref] [PubMed]
- Lin J, Fan X, Chen J, et al. Small interfering RNA-mediated knockdown of KRT80 suppresses colorectal cancer proliferation. Exp Ther Med 2020;20:176. [Crossref] [PubMed]
- Song H, Xu Y, Xu T, et al. CircPIP5K1A activates KRT80 and PI3K/AKT pathway to promote gastric cancer development through sponging miR-671-5p. Biomed Pharmacother 2020;126:109941 [Crossref] [PubMed]
- Morgan HJ, Benketah A, Olivero C, et al. Hair follicle differentiation-specific keratin expression in human basal cell carcinoma. Clin Exp Dermatol 2020;45:417-25. [Crossref] [PubMed]
- Rupertus K, Sinistra J, Scheuer C, et al. Interaction of the chemokines I-TAC (CXCL11) and SDF-1 (CXCL12) in the regulation of tumor angiogenesis of colorectal cancer. Clin Exp Metastasis 2014;31:447-59. [Crossref] [PubMed]
- Gao YJ, Liu L, Li S, et al. Down-regulation of CXCL11 inhibits colorectal cancer cell growth and epithelial-mesenchymal transition. Onco Targets Ther 2018;11:7333-43. [Crossref] [PubMed]
- Eftang LL, Klajic J, Kristensen VN, et al. GFRA3 promoter methylation may be associated with decreased postoperative survival in gastric cancer. BMC Cancer 2016;16:225. [Crossref] [PubMed]
- Wu ZS, Pandey V, Wu WY, et al. Prognostic significance of the expression of GFRα1, GFRα3 and syndecan-3, proteins binding ARTEMIN, in mammary carcinoma. BMC Cancer 2013;13:34. [Crossref] [PubMed]
- Tang JZ, Kong XJ, Kang J, et al. Artemin-stimulated progression of human non-small cell lung carcinoma is mediated by BCL2. Mol Cancer Ther 2010;9:1697-708. [Crossref] [PubMed]
(English Language Editor: J. Teoh)



